Posters – PSC Symposium 2021
Find below all posters for the PSC Symposium 2021 Patterns in Nature and Plant Sciences to be held online on December 8th 2021.
Break-out Room 1 chaired by Cyril Zipfl
1_Abarca, Alicia_AtRALF peptides as versatiles signaling peptides
AtRALF peptides as versatiles signaling peptides
Alicia Abarca1, Christina M. Franck and Cyril Zipfel1,2
1Department of Plant and Microbial Biology and Zurich-Basel Plant Science Center, University of Zurich, 8008 Zurich, Switzerland.
2The Sainsbury Laboratory, University of East Anglia, Norwich Research Park, Norwich, NR4 7UH, United Kingdom.
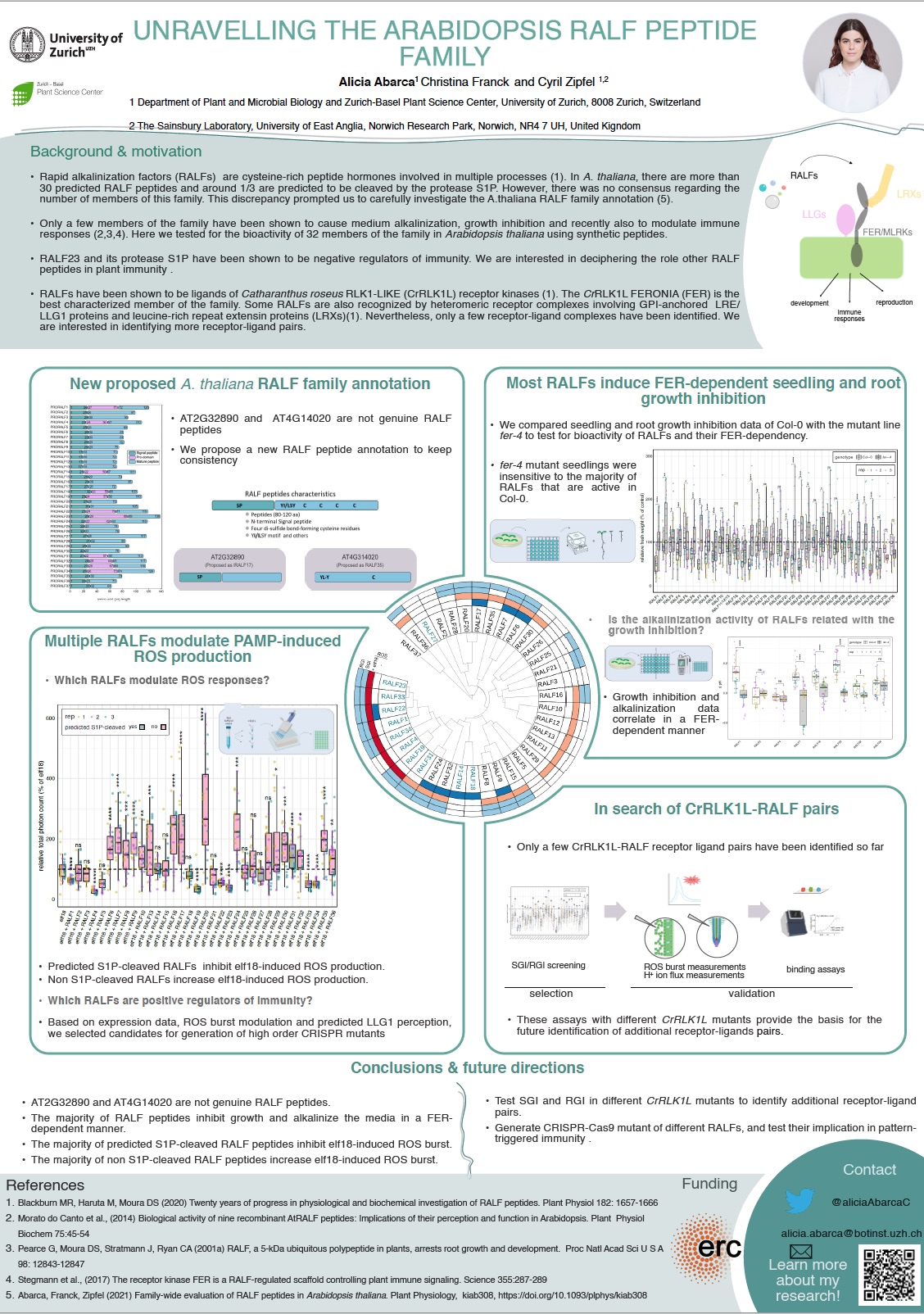
The cell wall-apoplast-plasma membrane axis represents a highly coordinated and dynamic barrier between the outside and the inside of the plant that efficiently perceive and respond to environmental cues. Nevertheless, plant innate immunity is energetically expensive and represent a growth-trade-off. Therefore, it has to be tightly regulated in order to be economic and efficient to the plant. Recently, our laboratory identified that a small peptide from the RAPID ALKALINIZATION FACTOR (RALF) family is a negative regulator of immunity. It inhibits ligand-induced immune receptor complex formation and decreases the levels of ROS production after elicitor perception. However, to date, there was no comprehensive, family-wide functional study on RALF peptides. Our phylogenetic analysis revealed that two of the previously proposed RALF peptides are not genuine RALF peptides, which lead us to propose a new consensus AtRALF peptide family annotation. We also showed that the majority of AtRALF peptides are able to induce seedling or root growth inhibition in A. thaliana seedlings when applied exogenously as synthetic peptides. Moreover, our findings suggest that alkalinization and growth inhibition are mostly coupled characteristics of RALF peptides and that all these responses are dependent on the Catharanthus roseus RLK1-LIKE receptor kinase FERONIA for the majority of the peptides, suggesting a pivotal role in the perception of multiple RALF peptides. Additionally, we identified additional RALF family members that negatively modulate immunity and conversely we identified RALF peptides that play a positive role in the regulation of ROS responses induced by elicitors. Future genetical and biochemical investigations will allow us to further understand which peptides of this family play a role as phytocytokines in A. thaliana and how this is orchestrated mechanistically.
2_Akiyma, Reiko_Time-course image analysis of Arabidopsis thaliana and its wild relatives in fluctuating field conditions using machine learning
Time-course image analysis of Arabidopsis thaliana and its wild relatives in fluctuating field conditions using machine learning
Reiko Akiyma1, Takao Goto2, Toshiaki Tameshige3 , Jiro Sugisaka3,4, Rie Shimizu-Inatsugi1, Jianqiang Sun5, Junichi Akita6, Natsumaro Kutsuna2, Jun Sese7,8,9, Kentaro K. Shimizu1,3
1 Department of Evolutionary Biology and Environmental Studies, University of Zurich
2 Research and Development Division, LPixel Inc.
3 Kihara Institute for Biological Research (KIBR), Yokohama City University
4 Center for Ecological Research, Kyoto University
5 Research Center for Agricultural Information Technology, National Agriculture and Food Research Organization (NARO), Japan
6 Department of Electric and Computer Engineering, Kanazawa University
7 Humanome Lab, Inc.
8 Artificial Intelligence Research Center, AIST
9 AIST-Tokyo Tech RWBC-OIL
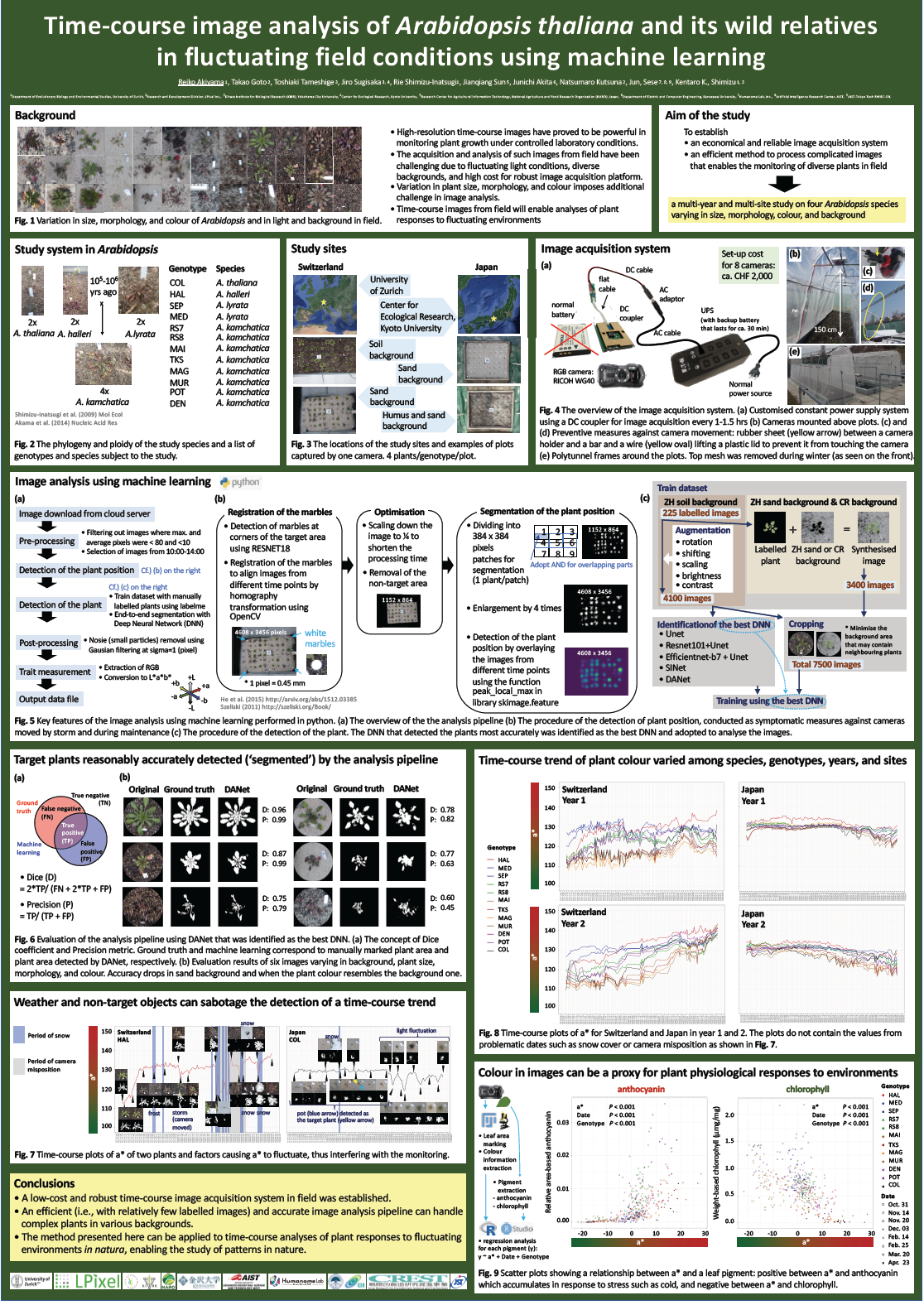
High-resolution time-course images has proved to be powerful in monitoring plant growth under controlled conditions. The acquisition and analysis of such images from field have been challenging due to fluctuating light conditions and diverse backgrounds, as well as high cost for robust image acquisition platform. To seek for an economical and reliable image acquisition system and an efficient method to process complicated images from field, we conducted a multi-year and multi-site study on four Arabidopsis species varying in size, morphology, colour, and background. We collected images daily for five months using commercial digital RGB cameras with a customised water shield and a battery system for constant power supply. The acquired images were analysed using a machine learning pipeline, where 225 plant images from soil background were labelled (1) to be augmented to yield training data for soil background, and (2) to be synthesised into sand and humus backgrounds to yield training data for these backgrounds. Using these training data, we performed an end-to-end segmentation with deep neural networks to identify plants in the image. The pipeline accurately recognised the target plants, from which we extracted colour information. Time-course colour trend varied among species, years, and sites. Quantification of leaf pigments indicated that the colour of the study species correlated with the amount of anthocyanin and chlorophyll, suggesting that colour information extracted from RGB images is effective as a proxy for plant physiological response to the environment. In summary, we have established a low-cost and robust time-course image acquisition system in field and an efficient (i.e., with relatively few labelled images) and accurate image analysis pipeline that can handle complex plants in various backgrounds. The method presented here can be applied to time-course analyses of plant responses to fluctuating environments in natura.
3_Baan, Jochem_Phylogenetic patterns in leaf wax n-alkane hydrogen isotope composition can be observed in spatially separated parts of the biosynthetic pathway
Phylogenetic patterns in leaf wax n-alkane hydrogen isotope composition can be observed in spatially separated parts of the biosynthetic pathway
Jochem Baan1, Meisha Holloway-Phillips1, Jurriaan M. de Vos1, Daniel B. Nelson1, Ansgar Kahmen1
1 University of Basel, Department of Environmental Sciences - Botany
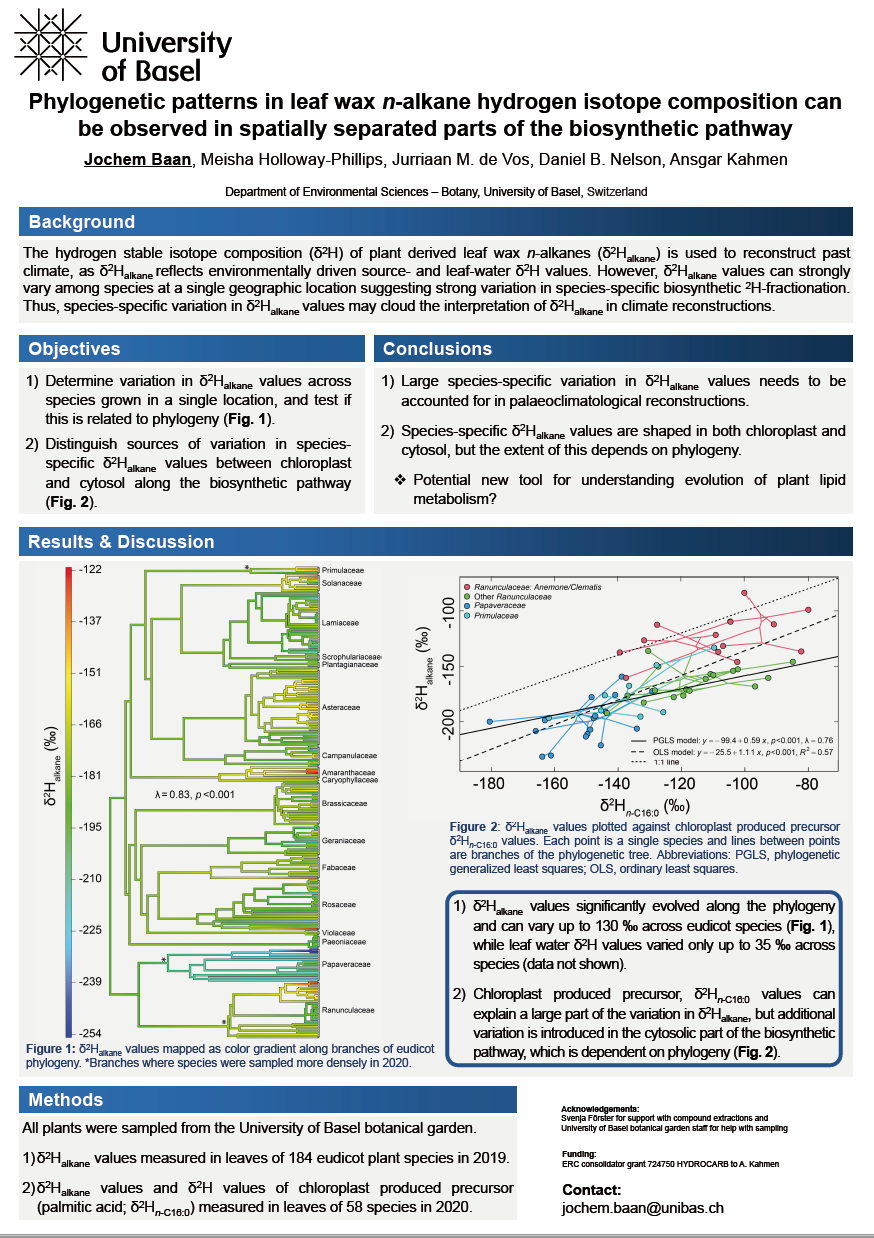
Hydrogen stable isotope ratio (δ2H) analyses of plant derived n‑alkanes have been developed as tools for ecological, environmental and palaeoclimatological studies, as the hydrogen stable isotope ratio of n‑alkanes (δ2Halkane) records source and leaf water δ2H values. However, large variation in δ2Halkane values among species at a single geographic location has been observed which suggests strong variation in species-specific biosynthetic 2H-fractionation. As such, the interpretation of δ2Halkane values in climate reconstructions is clouded by species-specific variation in δ2Halkane values. To explore variation in δ2Halkane values among species, we measured δ2Halkane values in a total of 184 eudicot plant species grown in a single location in 2019, and test if phylogenetic relatedness structures variation in δ2Halkane values. Our results show that species-specific δ2Halkane values can vary upwards of 130 ‰, and species variation in δ2Halkane significantly evolved along the phylogeny. This highlights the necessity of accounting for species-specific variation in δ2Halkane values when used for palaeoclimatological reconstructions. To get a better understanding of possible mechanisms that cause variation in species-specific biosynthetic 2H-fractionation, in a separate sample set, we aimed at distinguishing sources of variation in δ2Halkane values in different parts of the biosynthetic pathway. The acetogenic pathway, in which n-alkanes are synthesized, is spatially separated within cells between the plastid and cytosol. Therefore, we specifically measured δ2Halkane values and compared these to δ2H values of chloroplast‑produced precursors (palmitic acid; δ2Hn‑C16:0) of 58 eudicot plant species grown in a single location in 2020. Values of δ2Hn‑C16:0 and δ2Halkane showed a significant positive correlation, suggesting that variation in δ2Halkane values is largely shaped by processes in the plastid leading up to the synthesis of palmitic acid. However, species-specific variation in the isotopic offset between δ2Hn‑C16:0 and δ2Halkane values, which represents variation in δ2H values that occurs in cytosolic part of the biosynthetic pathway, was also related to phylogeny. This illustrates that the extent to which the two spatially separated parts of the acetogenic pathway influence δ2Halkane depends on species identity due to phylogenetic effects.
4_Bernasconi, Alessio_The role of the plant immune system in a virulent-avirulent strain coinfection
The role of the plant immune system in a virulent-avirulent strain coinfection
Alessio Bernasconi1, Bruce McDonald1, Andrea Sánchez-Vallet1
1 Plant Pathology, Institute of Integrative Biology (IBZ),ETH Zurich
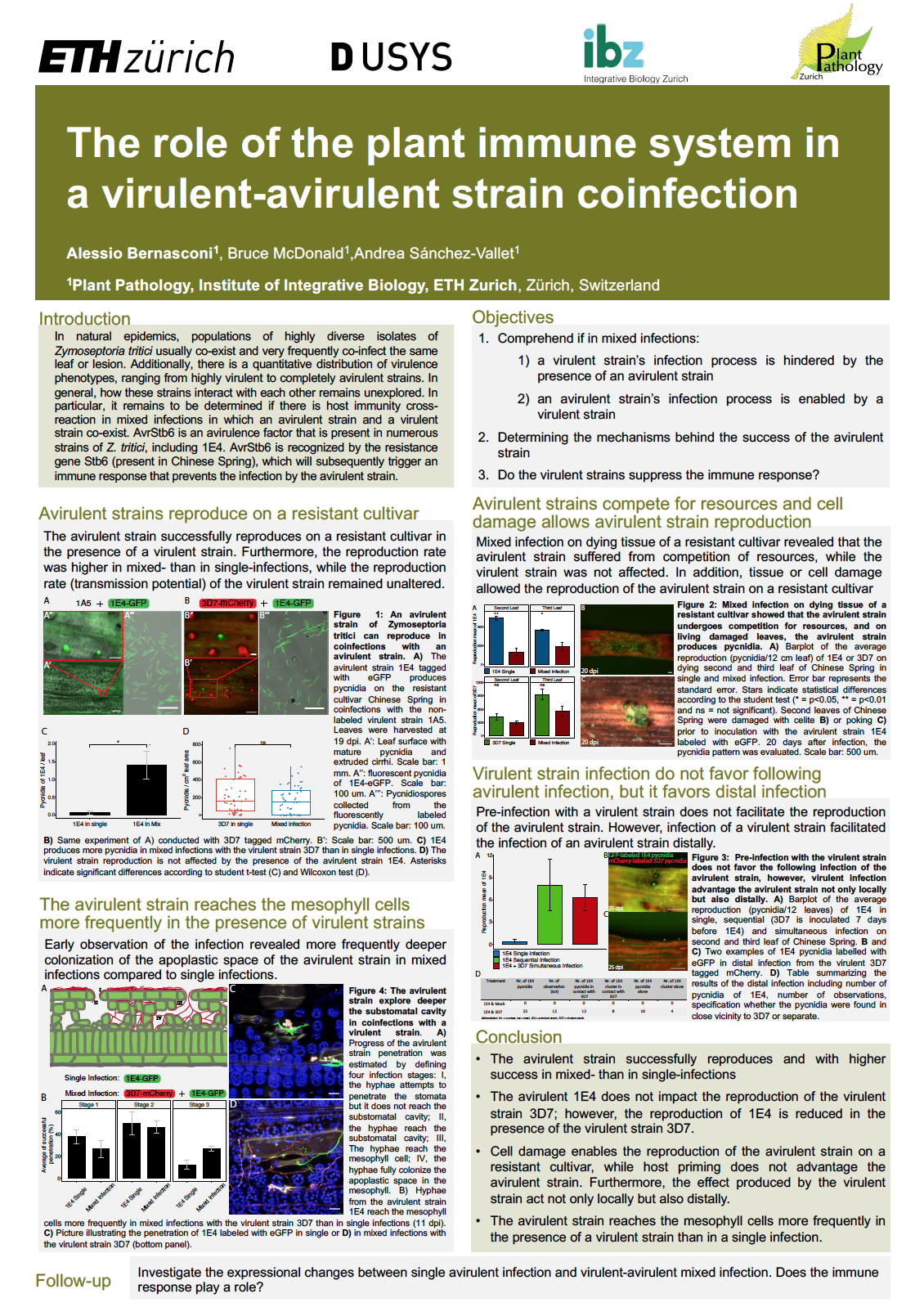
Following the gene-for-gene interaction, a strain that carries an avirulent gene is recognized by the corresponding resistant gene on the plant counterpart leading to an incompatible reaction. Consequently, in a single plant-pathogen system, the avirulent strain of, for example, Zymoseptoria tritici is unable to infect and reproduce on that specific cultivar, such as Chinese Spring. However, natural infections are frequently produced by several strains that co-infect simultaneously the same host. How the avirulent strain interacts with a resistant cultivar in presence of a virulent strain in mixed infection remains poorly understood. Recently, we demonstrated the substantial advantage offered by the mixed infection that significantly increases the opportunity of asexual reproduction of the avirulent strain on a resistant cultivar. We additionally showed that tissue or cell damage was sufficient for penetration and the reproduction of the avirulent strain. Nevertheless, in distal infection, the avirulent strain seems to profit from the same advantage, and confocal observation revealed early colonization of the plant tissue by the avirulent strain in co-infection with a virulent strain. We suggest that the repression of the immune system response caused by the virulent strain prevents avirulent strain recognition on a resistant cultivar.
5_Bernasconi, Zoe_Unravelling the molecular basis of wheat powdery mildew’s virulence patterns through ultraviolet mutagenesis
Unravelling the molecular basis of wheat powdery mildew’s virulence patterns through ultraviolet mutagenesis
Zoe Bernasconi1, Ursin Stirnemann1, Javier Sánchez-Martín1
1 Department of Plant and Microbial Biology, University of Zurich,
Resistance (R) genes play a major role in plant immunity, conferring race-specific resistance to pathogens. R genes can recognize avirulence (Avr) effectors that fungal pathogens secrete during infection. However, pathogens deploy various mechanisms to overcome this resistance, such as mutations, reduced expression of Avrs, or the presence of suppressors of recognition. Therefore, a complex set of multiple R/Avr interactions determines the final resistance or susceptibility of a host plant.
In the wheat (Triticum aestivum) - powdery mildew (Blumeria graminis f. s. tritici, Bgt) pathosystem, many R/Avr gene pairs have been characterized. The Pm3 allelic series is one of the most extensively studied. Different Pm3 isoforms, such as Pm3a or Pm3b, specifically recognize unrelated Avrs from Bgt (AvrPm3a/f and AvrPm3b/c respectively), thus leading to resistance. However, in some cases the pathogen’s virulence pattern cannot be solely explained by the Pm3/AvrPm3 interaction alone, indicating that other virulence determinants remain unknown.
We developed an ultraviolet (UV) mutagenesis-based approach to identify and characterize genes affecting virulence in Bgt. We obtained 24 Bgt mutants with a gain of virulence on different Pm3 containing wheat lines. Surprisingly, most of the Pm3 virulent Bgt mutants did not have mutations in the genes that were previously shown to be involved in virulence (e.g. AvrPm3). Moreover, qRT-PCR experiments showed that the expression of some Avrs in the virulent Bgt mutants was altered. This indicates the presence of novel genetic components that can modulate gene expression, thus causing race-specific gain of virulence.
UV mutagenesis has the potential to deepen our understanding of virulence patterns of wheat powdery mildew. This knowledge will enable us to develop diagnostic tools for the monitoring and the control of emerging virulent races of this major wheat pathogen.
6_Danli, Fei_Plant reproduction: chromatin-based controls in the reproductive lineage
Plant reproduction: chromatin-based controls in the reproductive lineage
Danli Fei1, Célia Baroux1
1 Department of Plant and Microbial Biology, University of Zurich
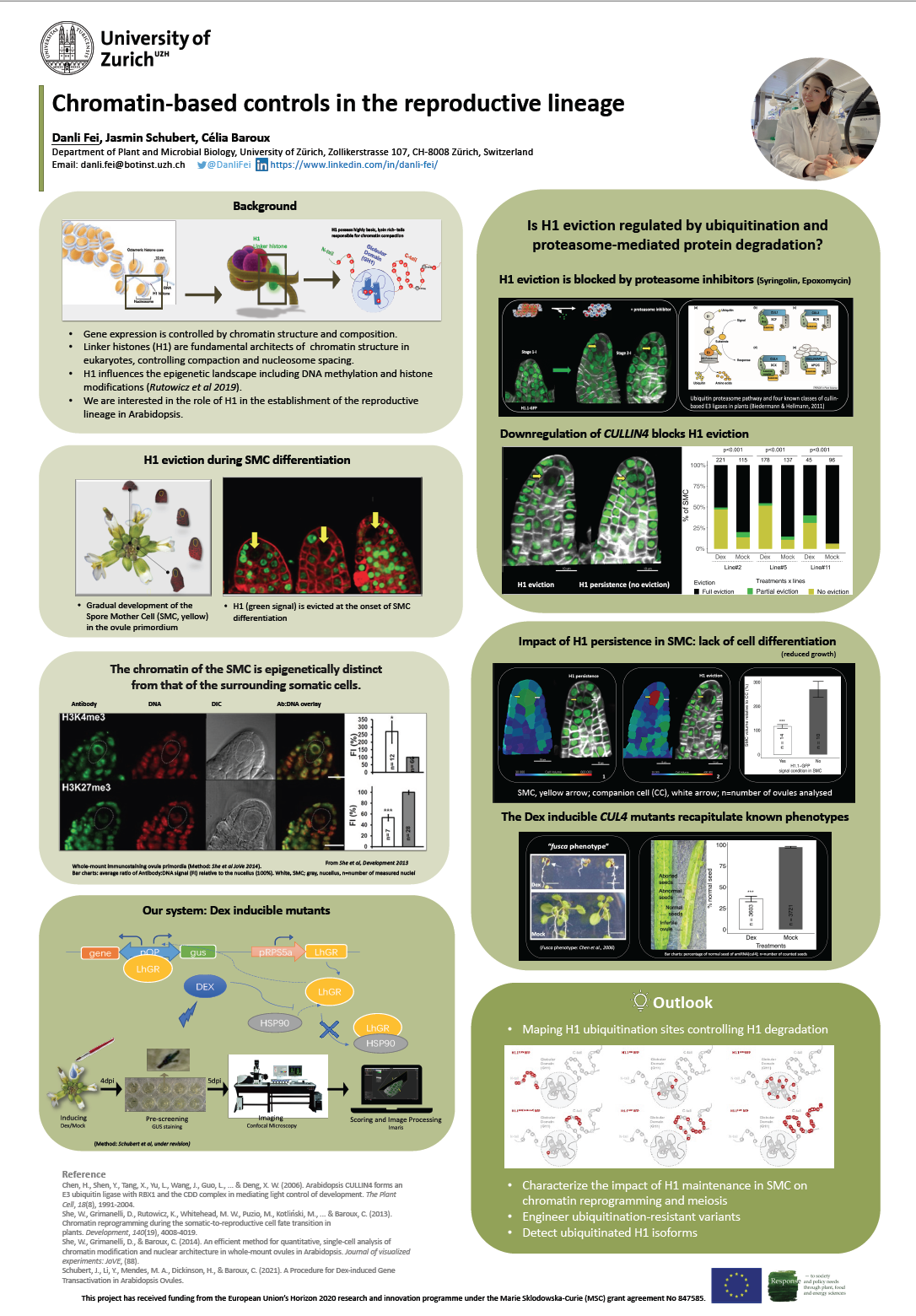
The nucleus is more than a genetic container. This organelle is the chief orchestra of cellular processes by controlling and fine-tuning gene expression in response to developmental and environmental cues. The combination of DNA and histone proteins that make up the nuclear content is often referred to as chromatin. The function of chromatin is packaging long DNA molecules into more compact, denser structures. Linker histone is the key composition of chromatin, which binds the nucleosome at the entry and exit sites of the DNA. The modification of this structural proteins in chromatin alters local chromatin structure and therefore gene expression.
Our research group aims to elucidate chromatin dynamics principles underlying cellular reprogramming during developmental or physiological transitions. My project focuses on the somatic-to-reproductive cell fate transition which leads to germline differentiation and then seed formation. Our group has shown that, in the model plant Arabidopsis, the differentiation of both male and female spore mother cells (SMC) is accompanied by large-scale chromatin reprogramming including the loss of linker histones (H1), chromatin decondensation, and large-sclae epigenetic changes (She et al., 2013, 2015). Specifically, the goal of my project is to address the role and mechanisms of H1 dynamics during female sporogenesis in Arabidopsis, focusing on ubiquitinylation and the proteasome-degradation pathway. The purpose of this research is to contribute knowledge on the molecular and epigenetic mechanisms controlling plant reproduction, in turn influencing seed yield.
To analyse H1 dynamics during female sporogenesis, we make use of engineered, inducible mutants. Some mutants downregulate specific components of the ubiquitination (Ub) pathway (CUL4) in the SMC while others ectopically express H1 mutant variants modified at candidate regions, potentially target sites of Ub. We combine methods in molecular biology (cloning, genotyping, DNA and RNA work, gene expression analyses), cell biology (nuclei isolation, tissue fixation and staining, immunolabeling) and microscopy imaging (light microscopy, fluorescence confocal microscopy) to describe chromatin organization at a microscopic and quantitative scale. Protocols for 3D quantitative analyses of chromatin composition and organization at the single-cell level in whole-mount plant tissues were established by our research group.
Break-out Room 2 chaired by Anne Roulin
7_Heutinck, Arvid_Branched malto-oligosaccharides cause spontaneous starch granule initiation in A. thaliana chloroplasts
Branched malto-oligosaccharides cause spontaneous starch granule initiation in A. thaliana chloroplasts
Arvid Heutinck1, Selina Camenisch1, Samuel C. Zeeman1
1 Plant Biochemistry, Institute of Molecular Plant Biology, ETH Zurich
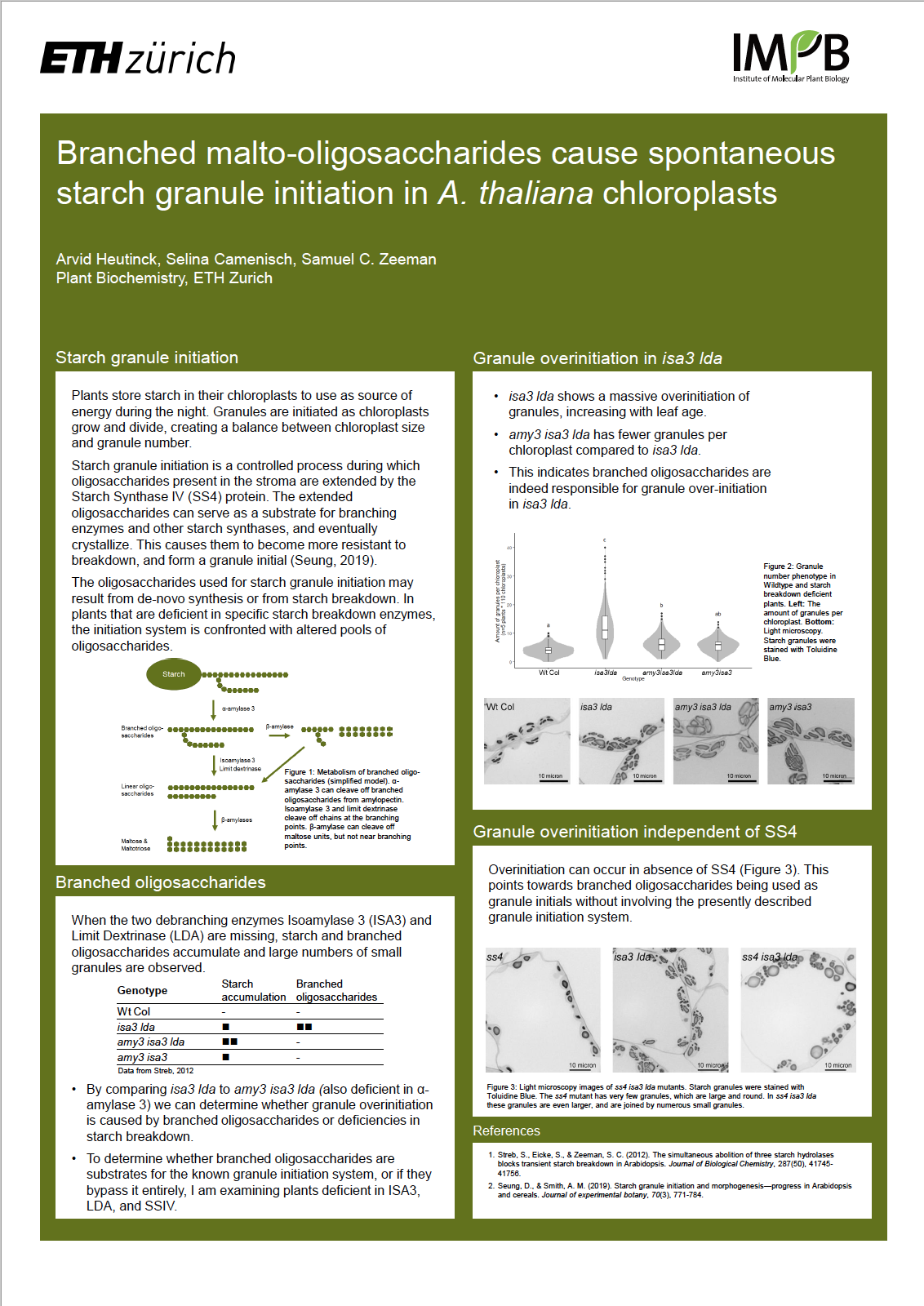
Plants store starch in their chloroplasts to use as source of energy during the night. A mature Arabidopsis chloroplast holds about 4-7 starch granules which grow during the day and shrink at night. Granules are initiated as chloroplasts grow and divide, creating a balance between chloroplast size and granule number.
Starch granule initiation is a controlled process during which oligosaccharides present in the stroma are extended by the Starch Synthase IV (SSIV) protein. The extended oligosaccharides can serve as a substrate for branching enzymes and other starch synthases, and eventually crystallize. This causes them to become more resistant to breakdown, and form a granule initial. Several proteins are involved in this process, and plants deficient in these show a decreased granule number. The oligosaccharides used for starch granule initiation may result from de-novo synthesis or from starch breakdown. In plants that are deficient in specific starch breakdown enzymes, the initiation system is confronted with altered pools of oligosaccharides.
When the two debranching enzymes Isoamylase 3 (ISA3) and Limit Dextrinase (LDA) are missing, branched oligosaccharides accumulate and large numbers of small granules are observed. We hypothesized that this over-initiation effect could be caused by the accumulation of branched oligosaccharides. To test this, I investigated the triple mutant isa3 lda amy3, which is also deficient in alpha-amylase 3 – the enzyme that can release branched oligosaccharides from starch. Compared to isa3 lda, the triple mutant has increased accumulation of starch, but no accumulation of branched oligosaccharides, and far fewer granules per chloroplast. This indicates branched oligosaccharides are indeed responsible for granule over-initiation in isa3 lda.
To determine whether branched oligosaccharides are substrates for the known granule initiation system, or if they bypass it entirely, I am examining plants deficient in ISA3, LDA, and SSIV. Preliminary data shows that despite the absence of SSIV, over-initiation still occurs. This points towards branched oligosaccharides being used as granule initials without involving the presently described granule initiation system. These findings give insight into how starch granules are initiated and established, and how this process can be influenced to initiate more.
8_Hou, Xiaoyu_Elucidating extensin-less LRX1-mediated dominant negative effect on cell wall development
Elucidating extensin-less LRX1-mediated dominant negative effect on cell wall development
Xiaoyu Hou1, Amandine Guerin1, Aline Herger1, Christoph Ringli1
1 Institute of Plant and Microbial Biology, University of Zurich
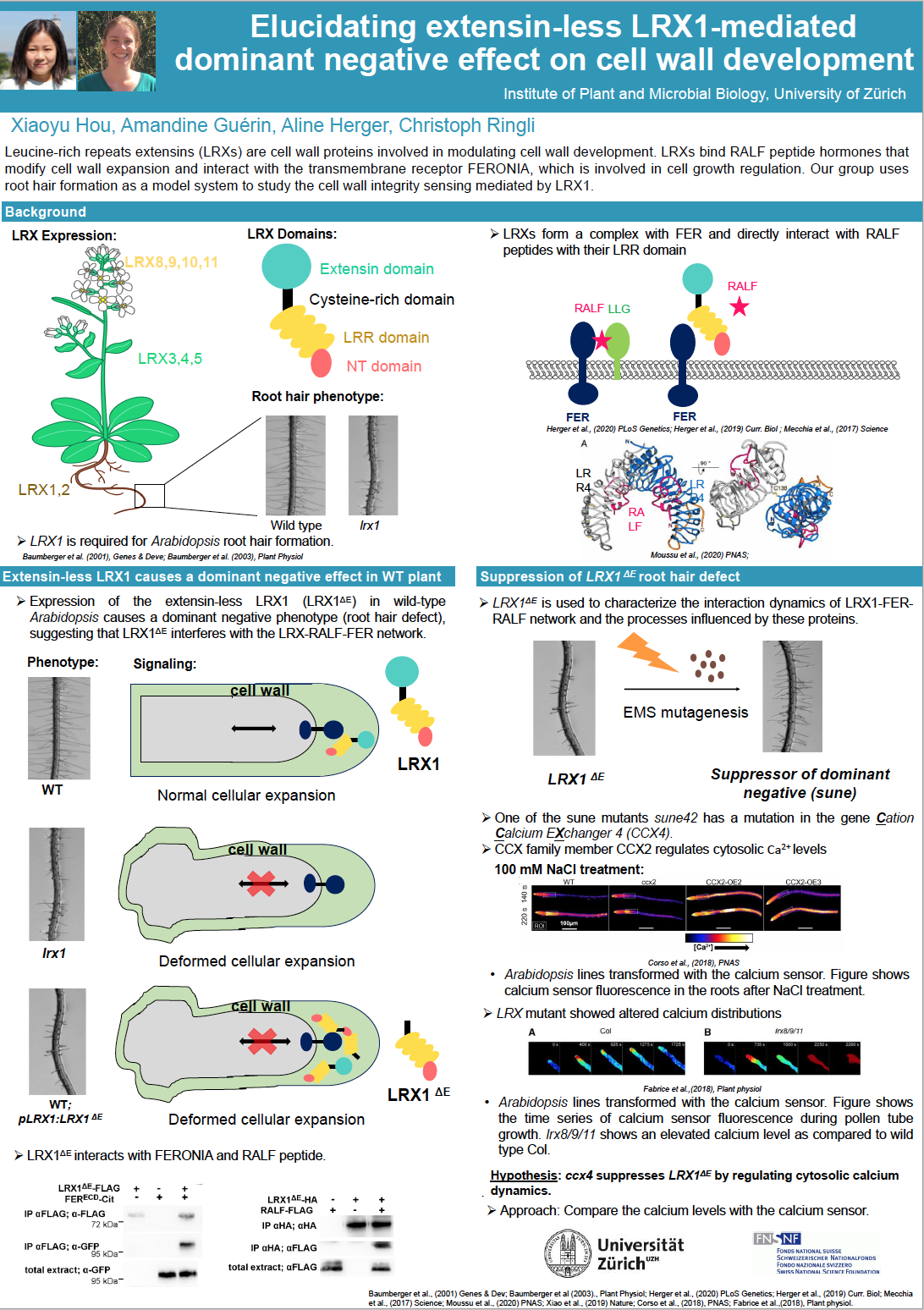
Plant cell growth requires the coordination between cell wall enlargement and protoplast expansion. To this end, plants have evolved an elaborate system to monitor cell wall homeostasis and convey the signals to intracellular signal cascades. The transmembrane protein kinase FERONIA (FER) functions in cell wall integrity sensing. It is a receptor of RALF (RAPID ALKALINIZATION FACTOR) peptide hormones that modulate plant cell growth. The extracellular proteins leucine-rich repeat extensins (LRXs) are identified as regulators for cell wall development. LRXs are high-affinity binding sites for RALF peptides. Interaction between FER and LRXs has been detected by protein-protein interaction assays. Therefore, our group proposed an LRX-RALF-FER module that regulates cell wall development.
The Arabidopsis genome encodes eleven members of LRXs. LRX8-11 are expressed in reproductive tissue, and LRX1-7 are expressed in vegetative tissue in which LRX1 and LRX2 are predominantly expressed in root hairs. Our group uses Arabidopsis root hairs as a model system to study the function of LRX1 in cell wall integrity. LRX1 and other LRXs possess an LRR domain for protein-protein interaction and an extensin domain which anchors the protein to the cell wall. Expression of the extensin-less LRX1 (LRX1ΔE) in wild-type Arabidopsis causes a dominant negative phenotype (root hair defect), suggesting that LRX1ΔE interferes with the LRX-RALF-FER network. Thus, this line is an excellent tool for characterizing the process that integrates intracellular signaling and cell wall integrity sensing
To investigate the effect of LRX1ΔE on LRX-RALF-FER interaction dynamics, we performed a genetic screen on LRX1ΔE. Suppression on LRX1ΔE- induced root hair defect was observed in several isolated mutants. Analysis on these mutants will gain us a better insight in the interaction dynamics of LRX-RALF-FER network. In addition, it will allow us to reveal physiological processes related to the cell wall integrity sensing.
9_Gonzales, Rhea_Strategies for improving forage productivity under future climates by genomics-assisted breeding
Strategies for improving forage productivity under future climates by genomics-assisted breeding
Reah Gonzales1,2, Steven Yates1, Chloe Manzanares1, Stéphane Charrier2 and Bruno Studer1
1 ETH Zurich, Institute of Agricultural Sciences, Molecular Plant Breeding,
2 Barenbrug SAS, France
Perennial ryegrass (Lolium perenne L.) and tall fescue (Festuca arundinacea Schreb.) are forage grasses widely grown in Europe. Cultivated forage grasses are affected by drought stress reducing biomass yield by up to 46% in perennial ryegrass and 20% in tall fescue. However, the reasons for different drought responses between the two species are unknown. In this project we investigate the physiological and genetic basis of drought tolerance in perennial ryegrass and tall fescue. The hypothesis being that drought induced reductions in plant productivity is quantifiable using high throughput phenotyping. To this end, we measure genotype-specific leaf elongation in response to drought using a novel high-throughput phenotyping platform. The system captures the specific soil moisture content at which leaf elongation reduces and stops. Moreover, the project links the high-throughput data with field data collected in a rain-out shelter, to test the utility of the lab-based phenotyping to predict crop performance in the real world. Finally, the plants will be genetically characterized using genotyping-by-sequencing, allowing us to test for marker trait associations. This will form the basis for marker development to assist breeding forage crops for future climates.
10_Grubinger, Thomas_Patterns of gene-tree variation leveraged from herbarium records reveal a complex evolutionary ancestry of early European tomatoes
Patterns of gene-tree variation leveraged from herbarium records reveal a complex evolutionary ancestry of early European tomatoes
Thomas Grubinger1, Gülfirde Akgül2, Alessia Guggisberg3, Reto Nyffeler4, Jurriaan M. de Vos5, Verena J. Schuenemann2, and Simon Aeschbacher1
1 Institute of Evolutionary Biology and Environmental Studies, University of Zürich
2 Institute of Evolutionary Medicine, University of Zürich
3 Department of Environmental Systems Science, ETH Zurich
4 Department of Systematic and Evolutionary Botany, University of Zürich
5 Department of Environmental Sciences – Botany, University of Basel
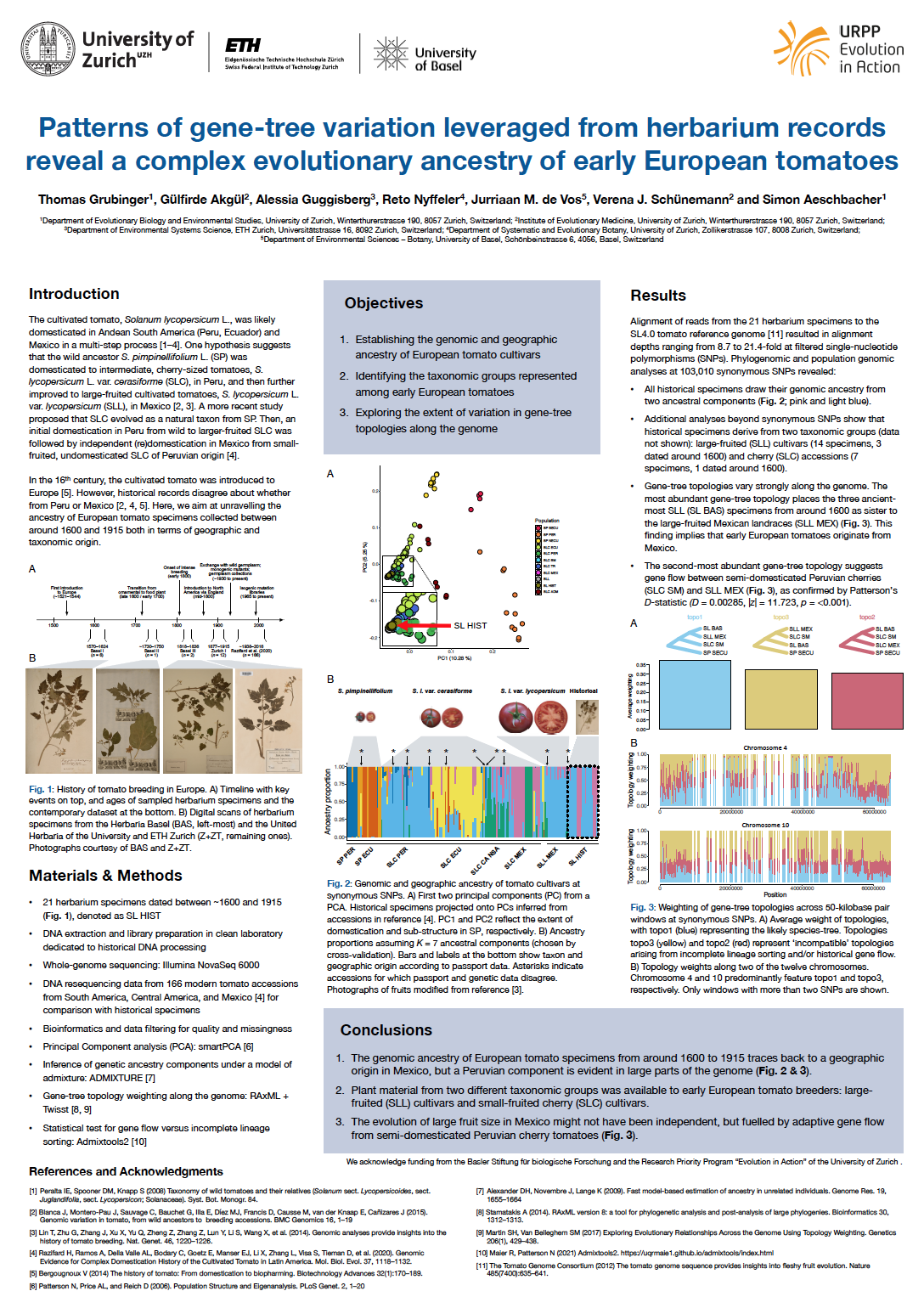
On its way to one of the most important vegetable crops today, the cultivated tomato (Solanum lycopersicum L.) was introduced to Europe in the 16th century. However, historical records disagree about whether early European tomatoes originated from Peru, Mexico or both. To address this question, we sequenced whole-genome ancient DNA from 21 herbarium specimens collected in Central Europe between 1596 and 1915. We combined these historical sequences with publicly available modern sequences from 166 wild, semi-domesticated, and landraDear Romyce accessions from Mexico, Central, and South America dated between 1938 and 1992. Our analyses of population structure and admixture at synonymous single-nucleotide polymorphisms (SNPs) revealed that all historical specimens draw their genomic ancestry from two components. One component fully represented in 5 historical specimens (2 from ~1600) is predominantly found in modern Mexican and Central American cherry tomatoes. The second ancestry component is fully represented in 10 historical specimens (3 from ~1600) and found in both large-fruited Mexican landraces as well as semi-domesticated Peruvian cherry tomatoes. The remaining 6 historical accessions (1 from ~1600) appeared to be admixed. While these results exclude a purely Peruvian origin of the earliest European tomatoes in our sample, the second ancestry component remained geographically inconclusive. To resolve the evolutionary history of this component, we inferred gene-tree topologies along the genome to quantify the support of competing evolutionary relationships among large-fruited Mexican landraces, semi-domesticated Peruvian cherry tomatoes, and the three oldest European specimens representing the geographically inconclusive ancestry component. Most gene-tree topologies (37.2%) suggested that the three historical specimens were sister to the large-fruited Mexican landraces, which suggests that our early European tomatoes all originated from Mexico. However, gene-tree topologies varied strongly along the genome. While both alternative species trees were about equally abundant on average, recombination rate and gene density appear to strongly drive relative gene-tree abundances. Our ongoing work aims at interpreting these patterns of short-scale genomic variation in evolutionary histories in terms of incomplete lineage sorting, admixture, and selection at known domestication genes underlying fruit morphology and flavour.
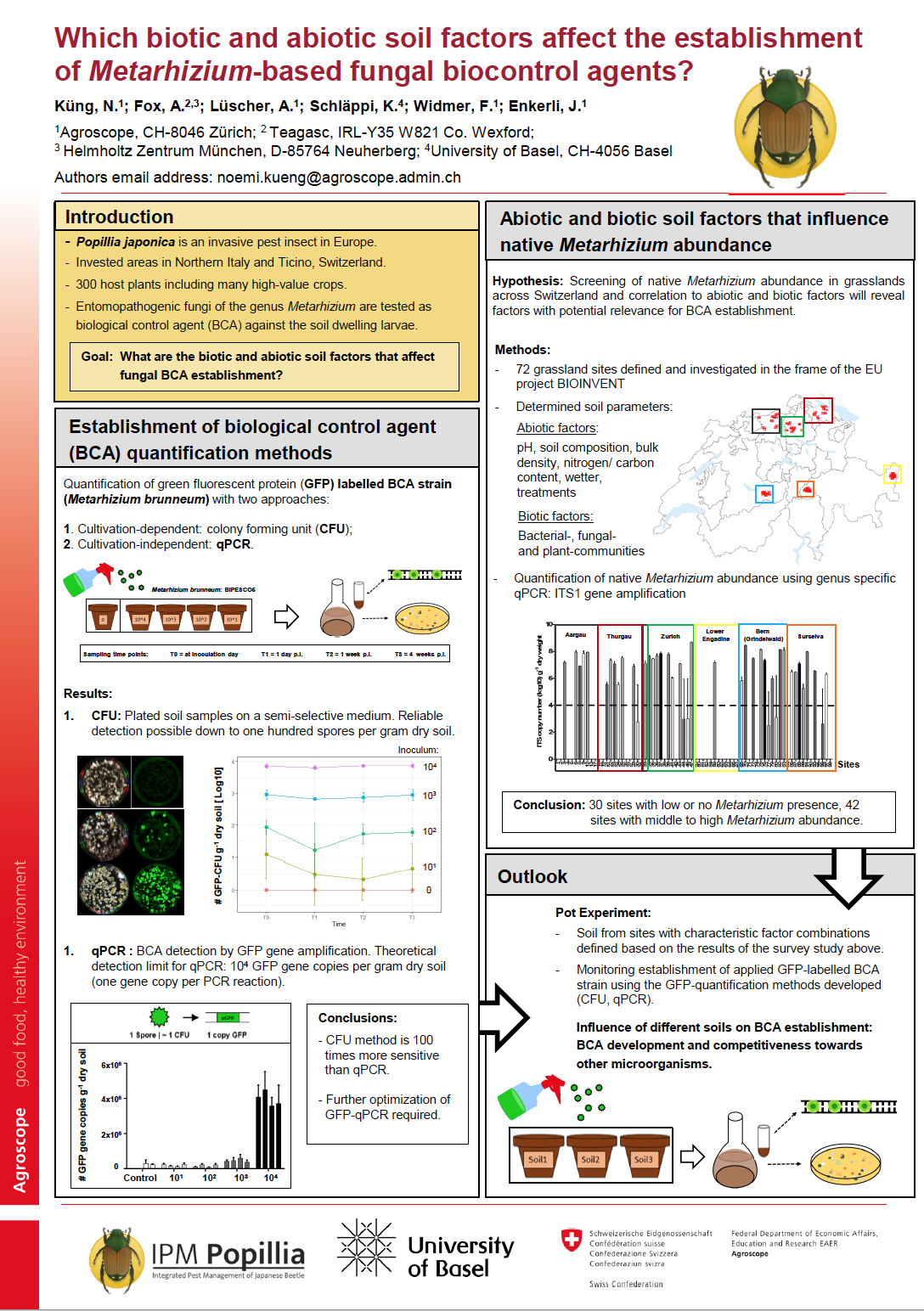
11_Küng, Noemi_Which biotic and abiotic soil factors affect the establishment of Metarhizium-based fungal biocontrol agents?
Which biotic and abiotic soil factors affect the establishment of Metarhizium-based fungal biocontrol agents?
Noemi Küng1, Aaron Fox2,3, Andreas Lüscher4, Klaus Schläppi5, Franco Widmer1, Jürg Enkerli1
1 Molecular Ecology, Agroscope, Zürich, Switzerland
2 Environment Research Centre, Teagasc, Johnstown Castle, Co. Wexford, Ireland
3 Research Unit Comparative Microbiome Analysis, Helmholtz Zentrum München, Munich, Germany
4 Forage Production and Grassland Systems, Agroscope, Zürich, Switzerland
5 Plant Microbe Interaction, University of Basel, Basel, Switzerland
Entomopathogenic fungal strains of the genus Metarhizium are commercially available and used as biocontrol agents (BCA) against a diverse range of insect pests. Because of their ability to infect and kill soil dwelling larvae, some strains have recently been tested as potential BCAs against the larvae of the invasive scarab beetle Popillia japonica, which currently infests regions in Northern Italy and Ticino, Switzerland. Beetles of this insect feed on up to three hundred different crops and cause significant economic damage.
From other well-established fungal BCAs, it is know that densities of up to 104 colony forming units (CFU) per gram soil are required to achieve reliable control of scarabid larvae in the soil. Potential drivers of fungal establishment include abiotic and biotic soil factors as well as presence of the host at sufficient density. The aim of this study is to decipher the soil factors, which drive fungal BCA establishment in the soil. First, GFP-based cultivation-dependent and cultivation-independent methods were established to provide a method that allows tracking of fungal development in soil using GFP-labelled Metharizium brunneum strains. The cultivation-dependent method, which is based on plating soil samples on a semi-selective medium and subsequent counting of fluorescent colony-forming units (CFU) has revealed a detection limit of one hundred CFUs per gram of soil. The cultivation-independent method, which is based on detection of GFP gene copy numbers by qPCR has revealed a detection limit of 104 gene copies per gram of soil. Second, the quantification methods will be used to monitor fungal BCA development and competitiveness towards other microorganisms in a pot experiment using different soils to investigate effects of the different soil factors, the native Metarhizium populations and the insect host presence. To allow selection of different soils for this experiment native Metarhizium abundance is currently being screened in samples obtained from 72 grassland sites in Switzerland, which were collected in a previous project (EU-project BIOINVENT). This study will provide in-depth information on biotic and abiotic soil factors that drive fungal BCA establishment and contribute to the establishment of a biological control strategy for the invasive pest P. japonica.
Break-out Room 3 chaired by Benjamin Stocker
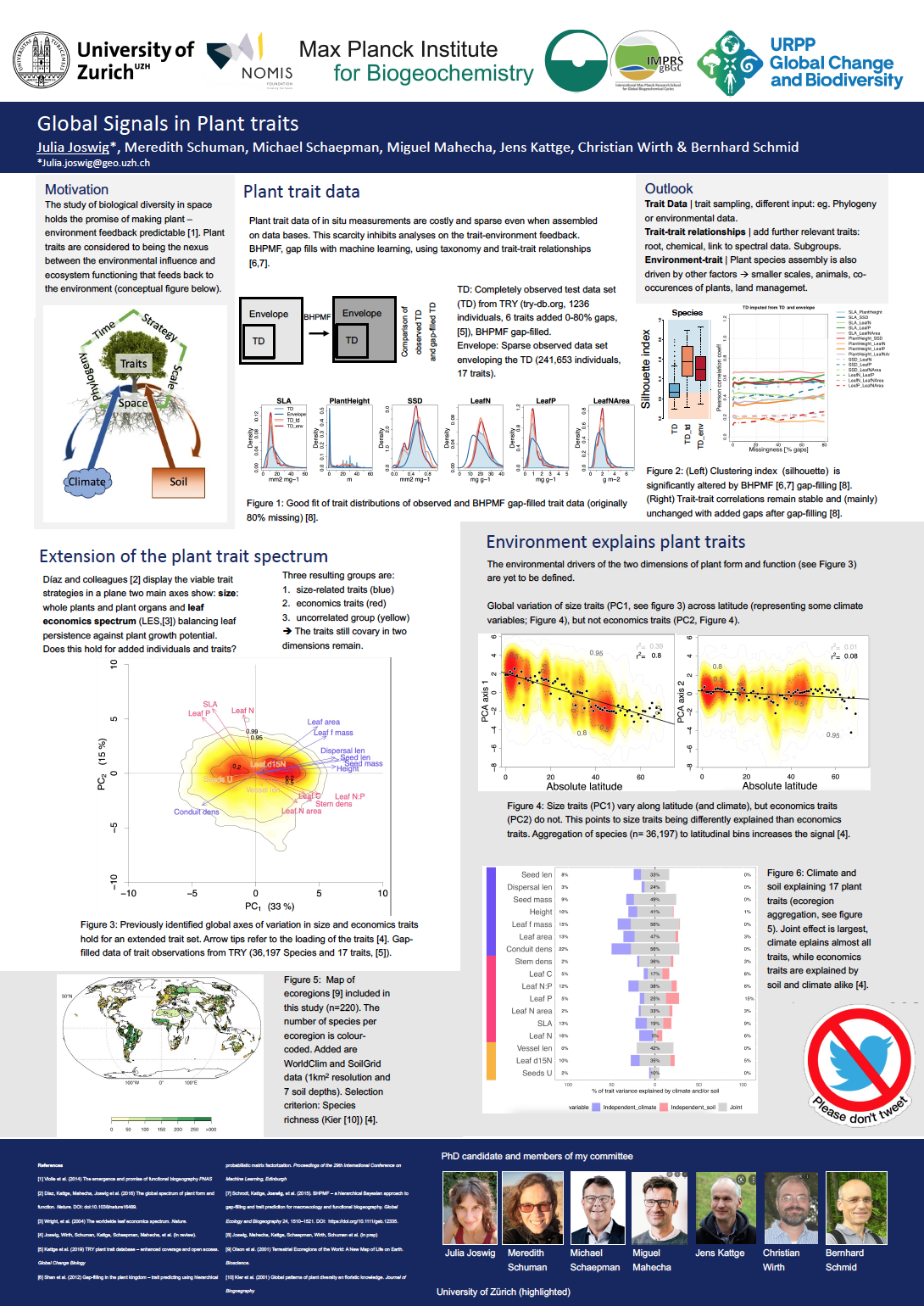
12_Joswig, Julia_Global Environmental Signals in Sparse Plant Traits
Global Environmental Signals in Sparse Plant Traits
Julia, Joswig1,2, Christian Wirth1,3,4, Meredith C. Schuman2,5, Jens Kattge1,3, Björn Reu6, Ian J. Wright7, Sebastian D. Sippel8,9, Nadja Rüger3,10,11, Ronny Richter3,4,12, Guido Kraemer1,3,50, Michael E. Schaepman2, Peter M. van Bodegom13, J. H. C. Cornelissen14, Sandra Dıaz15, Wesley N. Hattingh16, Koen Kramer17,18, Frederic Lens19,20, Ulo Niinemets21, Peter B. Reich22,23,24, Markus Reichstein1,3, Christine Römermann3,25, Franziska Schrodt26, Madhur Anand27, Michael Bahn28, Chaeho Byun29, Giandiego Campetella30, Bruno E. L. Cerabolini31, Joseph M. Craine32, Andres Gonzalez-Melo33, Alvaro G. Gutierrez34, Tianhua He35,36, Pedro Higuchi37, Herve Jactel38, Nathan J. B. Kraft39, Vanessa Minden40,41, Vladimir Onipchenko42, Josep Penuelas43,44, Valerio D. Pillar45, Enio Sosinski46, Nadejda A. Soudzilovskaia47,48, Evan Weiher49, Miguel D. Mahecha3,50,51
1 Max-Planck-Institute for Biogeochemistry, Jena, Germany
2 Remote Sensing Laboratories, Department of Geography, University of Zurich
3 German Centre for Integrative Biodiversity Research (iDiv) Halle-Jena-Leipzig
4 Institute of Systematic Botany and Functional Biodiversity, University of Leipzig, Germany
5 Department of Chemistry, University of Zurich
6 Escuela de Biología, Universidad Industrial de Santander, Bucaramanga, Colombia
7 Department of Biological Sciences, Macquarie University, Australia
8 Institute for Atmospheric and Climate Science, ETH Zurich
9 Norwegian Institute of Bioeconomy Research, Norway
10 Department of Economics, University of Leipzig, Germany
11 Smithsonian Tropical Research Institute, Apartado 0843-03092, Ancón, Panama
12 Geoinformatics and Remote Sensing, Institute for Geography, University of Leipzig, Germany
13 Environmental Biology Department, Institute of Environmental Sciences, CML, Leiden University, The Netherlands
14 Systems Ecology, Department of Ecological Science, Faculty of Science, Vrije Universiteit Amsterdam, The Netherlands
15 Instituto Multidisciplinario de Biología Vegetal (IMBIV), CONICET and FCEFyN, Universidad Nacional de Córdoba, Argentina
16 Global Systems and Analytics, Nova Pioneer, Johannesburg, South Africa
17 Chairgroup Forest Ecology and Forest Management, Wageningen University
18 Land Life Company, Mauritskade 63 1092AD Amsterdam, Netherlands
19 Naturalis Biodiversity Center, Research Group Functional Traits, The Netherlands
20 Leiden University, Institute of Biology Leiden, Plant Sciences,The Netherlands
21 Estonian University of Life Sciences,Tartu, Estonia
22 Department of Forest Resources, University of Minnesota, St Paul, USA
23 Hawkesbury Institute for the Environment, Western Sydney University, Australia
24 Institute for Global Change Biology and School for Environment and Sustainability, University of Michigan, Ann Arbor, Ml. USA
25 Institute of Ecology and Evolution, Dept. Plant Biodiversity, Friedrich-Schiller University Jena, Germany
26 School of Geography, University of Nottingham, University Park, Nottingham, UK
27 School of Environmental Sciences,University of Guelph, Canada
28 Department of Ecology, University of Innsbruck, Austria
29 Department of Biological Sciences and Biotechnology, Andong National University, Korea
30 School of Biosciences and Veterinary Medicine - Plant Diversity and Ecosystems Management Unit, University of Camerino, Italy
31 Departement of Biotechnologies and Life Sciences (DBSV)-University of Insubria, Varese, Italy
32 Jonah Ventures LLC, Boulder CO 80301, USA
33 Universidad del Rosario, Facultad de Ciencias Naturales y Matemáticas, Bogotá, Colombia
34 Departamento de Ciencias Ambientales y Recursos Naturales Renovables, Facultad de Ciencias Agronómicas, Universidad de Chile, Santiago, Chile
35 School of Molecular and Life Sciences, Curtin University, Perth, Australia
36 College of Science, Health, Engineering and Education, Murdoch University, Murdoch, WA, Australia
37 Department of Forestry, Universidade do Estado de Santa, Catarina, Lages, SC, Brazil
38 INRAE University Bordeaux, BIOGECO, F-33610 Cestas, France
39 Department of Ecology and Evolutionary Biology, University of California, 621 Charles E. Young Drive South, Los Angeles, USA
40 Department of Biology, Vrije Universiteit Brussel, Belgium
41 Landscape Ecology Group, Insitute of Biology and Environmental Sciences, University of Oldenburg, Germany
42 Department of Ecology and Plant Geography, Moscow State Lomonosov University, Russia
43 CSIC, Global Ecology Unit CREAF-CSIC-UAB, Bellaterra (Catalonia), Spain
44 CREAF, Cerdanyola del Vallès (Catalonia), Spain
45 Department of Ecology, Universidade Federal do Rio Grande do Sul, Porto Alegre, Brazil
46 Embrapa Recursos Genéticos e Biotecnologia, Brasília, Brazil
47 Centre for Environmental Sciences, Hasselt University, Belgium
48 Institute of Environmental Sciences, Leiden University, The Netherlands
49 Department of Biology, University of Wisconsin - Eau Claire, USA
50 Remote Sensing Centre for Earth System Research, University of Leipzig, Germany
51 Helmholtz Centre for Environmental Research, Leipzig, Germany
Plant functional traits can predict community assembly and ecosystem functioning and are thus widely used in global models of vegetation dynamics and land-climate feedbacks. Still, we lack a global understanding of how land and climate affect plant traits. Necessary plant trait data is sparse, but can be overcome by gap-filling. No bias control for gap-filled data exists beyond mere error estimation. In my PhD I present (1) how we overcome sparse data using gap-filling, evaluate any induced information and further analyse (2) how the previously found axes of trait variation, i.e. size and leaf economics are explained by environmental factors.
We (1) develop best practice for gap-filling of trait data and find that gap-filled data to be useful for analysing trait-trait relationships but not taxonomy. We (2) find the trait groups persist in a global dataset of 17 traits across more than 20,000 species. We find a dominant joint effect of climate and soil on trait variation. Additional independent climate effects are also observed across most traits, whereas independent soil effects are almost exclusively observed for economics traits. Variation in size traits correlates well with a latitudinal gradient related to water or energy limitation. In contrast, variation in economics traits is better explained by interactions of climate with soil fertility. These findings have the potential to improve our understanding of biodiversity patterns and our predictions of climate change impacts on biogeochemical cycles.
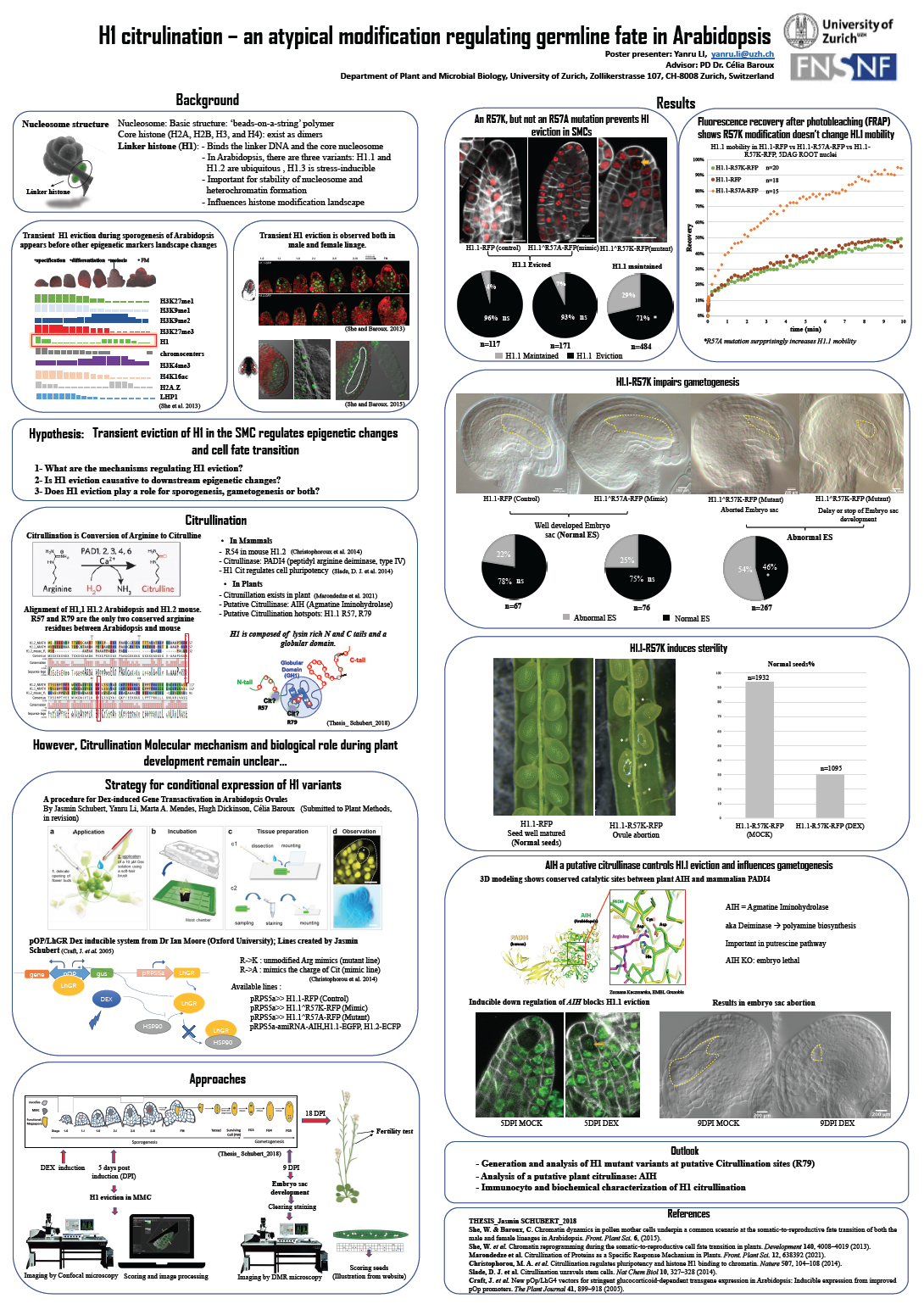
13_Li, Yanru_H1 citrulination – an atypical modification regulating germline fate in Arabidopsis
H1 citrulination – an atypical modification regulating germline fate in Arabidopsis
Yanru Li1, Célia Baroux1
1 Department of Plant and Microbial Biology, University of Zurich
Linker histones (H1) are essential architects of the 3D genome in eukaryotes and is a ubiquitous component of chromatin. Our group previously showed that in Arabidopsis, H1 undergoes transient eviction during the differentiation of spore mother cells (SMC), the plant’s germline precursors. Strikingly, H1 eviction precedes a broad range of large-scale chromatin changes affecting both structural organization and the epigenetic landscape. We hypothesise that H1 eviction plays a role in cell fate reprogramming during SMC differentiation, which marks the somatic-to-reproductive transition. To investigate this hypothesis, we first sought to identify the mechanisms controlling H1 eviction then to study the functional impact by altering this event. Based on homology with animal H1, we identified a conserved arginine in the Arabidopsis H1.1 variant (R57) playing an important role. Notably, an R57K but not an R57A mutation impairs H1 eviction in the SMC yet, without altering H1 properties in terms of stability on the chromatin. Failure in H1 eviction in the SMC led to post-meiotic defect including embryo sac abnormalities and reduced fertility. Furthermore, downregulation of an agmatine iminohydrolase (AIH), an enzyme is predicted to catalyze the conversion of arginine in citrulline, recapitulated these phenotypes. Collectively, these observations suggest H1 citrulination in the SMC as a key component regulating H1 eviction and germline fate.
14_Li, Cheng_Association study of genetic variation with variation in leaf reflectance
Association study of genetic variation with variation in leaf reflectance
Cheng Li1, Ewa A. Czyz1, Rayko Halitschke2, Ian T. Baldwin2, Michael E. Schaepman1, Meredith C. Schuman1
1 Department of Geography, University of Zurich
2 Department of Molecular Ecology, Max Planck Institute for Chemical Ecology
Plants are the trophic basis of terrestrial ecosystems and their diversity structures ecological communities. Genetic diversity is a key determinant of adaptive potential for species in a changing climate, and both plant genetic and species-level diversity has large effects on biodiversity experiments. Remote sensing of plants via the reflection of light from leaves and canopies facilitates large-scale and long-term repeat monitoring of plant populations in natural settings. How leaves reflect light is determined by aspects of physiology and function emerging from the interaction of plant genomes with the environment. These include leaf structure, and contents of pigments, water, and other abundant constituents like lignins, phenolics, and proteins.
We compared genetic and environmental influences on variance in leaf reflectance calculated from field spectroradiometer measurements using a standard light source and backgrounds. We used inbred lines of the wild coyote tobacco Nicotiana attenuata from wild accessions, recombinant inbred lines (RILs), and transgenic lines harboring targeted changes to gene expression. Plants were grown in more controlled (glasshouse) or more natural (field) environments.
We found that growth environment had a stronger effect than genetic differences on total variance in leaf reflectance, with least variance among plants grown in the glasshouse. Across all genotypes and environments, short-wave infrared (SWIR) regions influenced by leaf water and dry matter (e.g. protein, lignin, phenolics) contributed most to the first principle component of variance; the near-infrared (NIR), influenced by leaf structure, to the second; and the visible region, dominated by leaf pigments (VIS), to the third. Within environments, genetic variation specifically caused variation in leaf reflectance, even though time of day of measurement also caused reflectance differences within genotypes.
In summary, we describe variation in leaf reflectance resulting from genetic influences over specific physiological processes, accounting for environmental and temporal variation in leaf reflectance data. This supports the application of remote sensing to monitor genetic variation and related adaptation and physiological acclimation processes in plant populations.
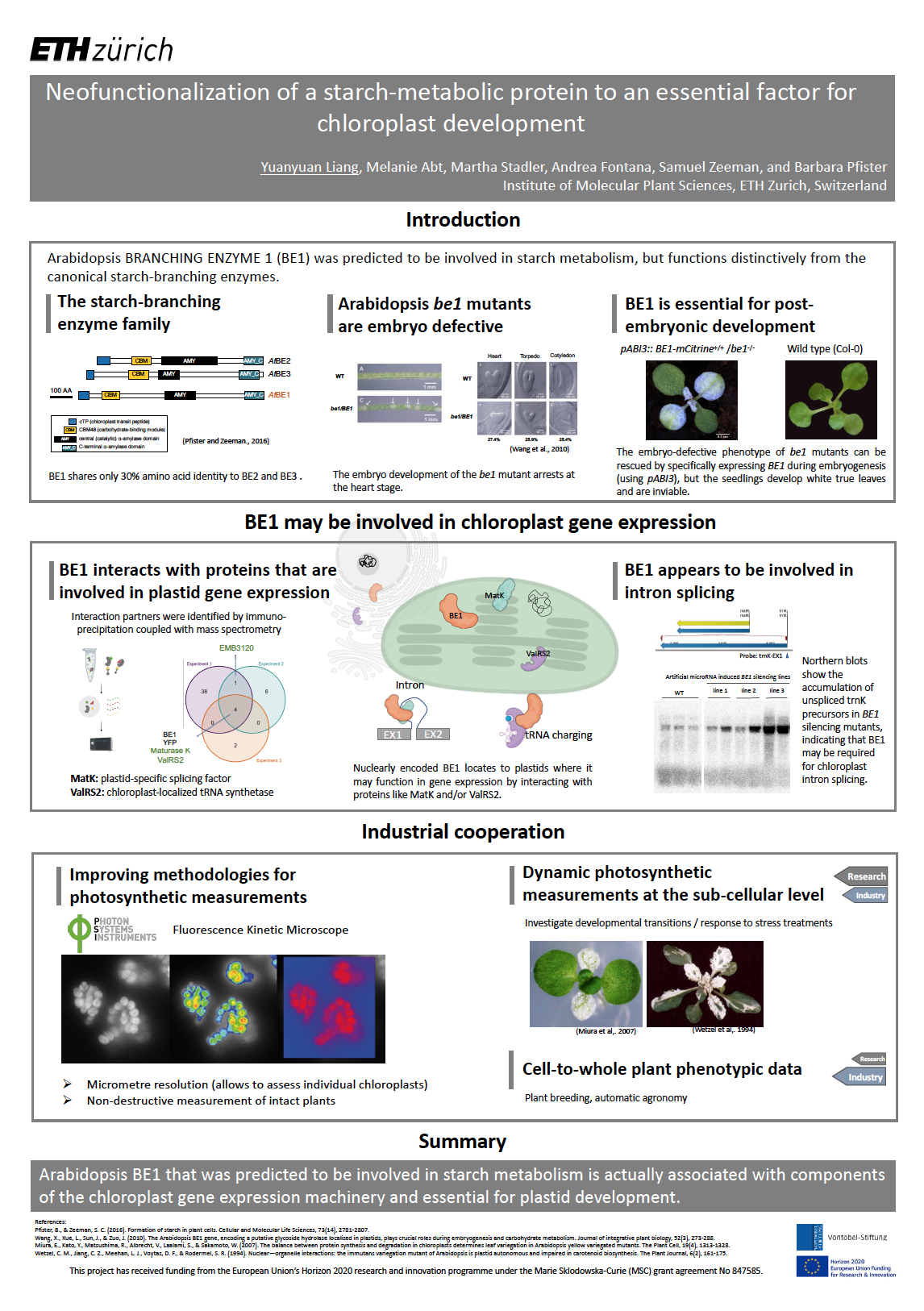
15_Liang, Yuanyuan_Neofunctionalization of a starchmetabolic protein to an essential factor for chloroplast development
Neofunctionalization of a starchmetabolic protein to an essential factor for chloroplast development
Yuanyuan Liang1, Melanie Abt1, M. Stadler1, A. Fontana1, S.C. Zeeman1, B. Pfister1
1 Institute of Molecular Plant Biology, ETH Zurich
Understanding the mechanisms by which chloroplasts develop and maintain their function under adverse conditions is of unparalleled global significance. As they house photosynthesis, even small alterations in chloroplast function imposed by environmental stresses can result in drastic yield losses. Maintaining crop yields in the face of increasingly unfavorable environmental conditions requires innovations that allow improvements in the plastid’s housekeeping metabolism and hence the stress resistance of these crucial organelles.
In this project, we aim to identify factors involved in the fundamental processes of chloroplast homeostasis. Specifically, our recent data suggests that an Arabidopsis
protein that was predicted to be involved in carbohydrate metabolism is actually associated with components of the plastid gene expression machinery and essential for plastid development. We discovered that this protein interacts with a chloroplastspecific splicing factor and a tRNA synthetase, both of which are essential plastidlocalized proteins. Silencing mutants of the underlying genes show an accumulation of precursors of chloroplast transcripts, which implies a role in chloroplast intron splicing and may explain their crucial functions in chloroplast development. Transcriptome analysis with a special focus on changes in chloroplast transcripts will be used to further investigate their impact on gene expression.
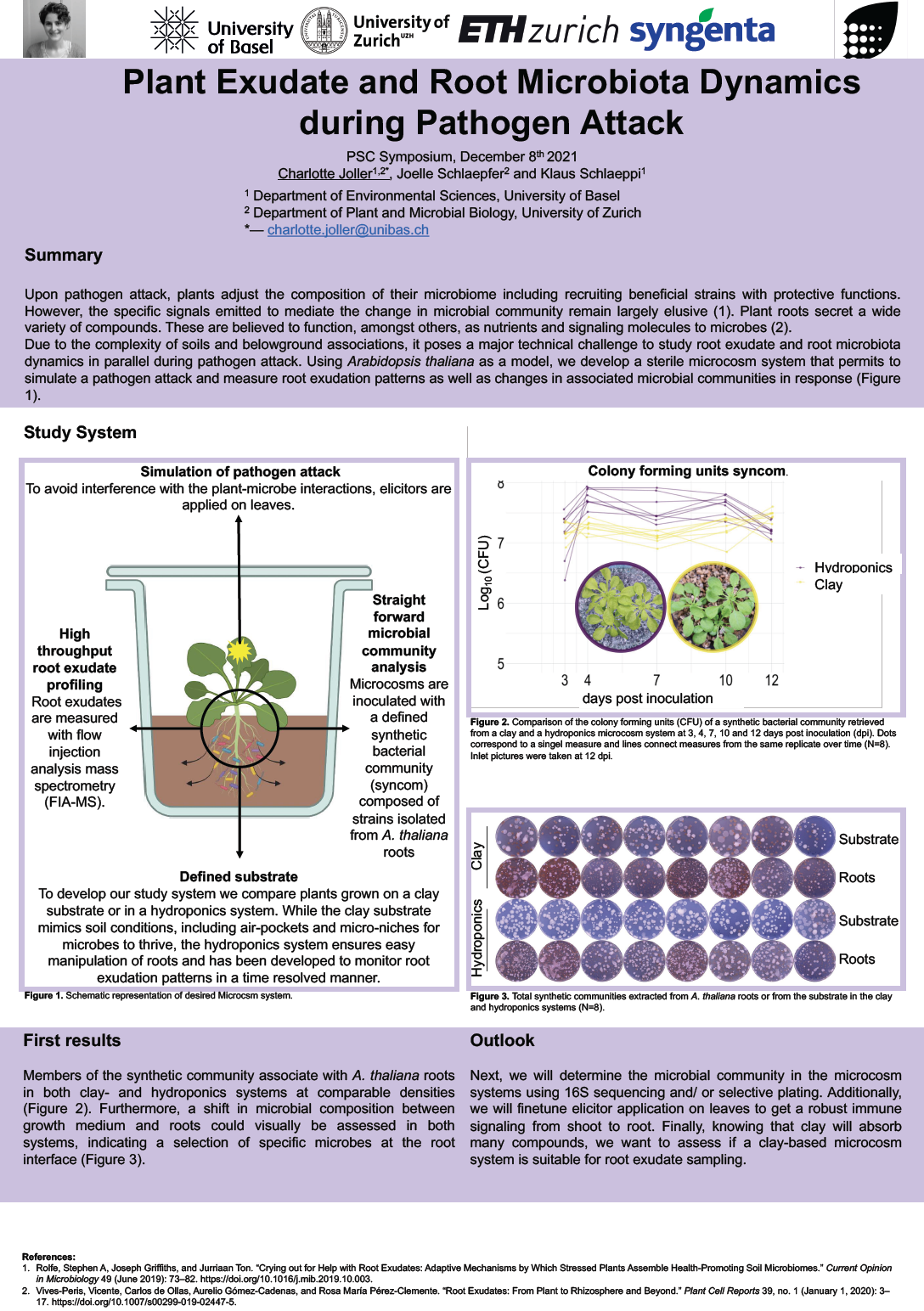
16_Joller, Charlotte_Plant Exudate and Root Microbiota Dynamics during Pathogen Attack
Plant Exudate and Root Microbiota Dynamics during Pathogen Attack
Charlotte Joller1,2, Joelle Schlaepfer2 and Klaus Schlaeppi1
1 Department of Environmental Sciences, University of Basel
2 Department of Plant and Microbial Biology, University of Zurich
Upon pathogen attack, plants adjust the composition of their microbiome. This includes recruiting of beneficial strains with protective functions. However, the specific signals emitted to mediate the change in microbial community remain largely elusive. Plant roots secret a wide variety of compounds. These are believed to function, amongst others, as nutrients and signaling molecules for microbes. We hypothesize that they present the mechanistic link between pathogen recognition and an altered composition of the plant-associated microbial community.
Due to the complexity of soils and belowground associations, it poses a major technical challenge to study root exudate and root microbiota dynamics in parallel during pathogen attack. Using Arabidopsis thaliana as a model, we develop a sterile microcosm system that permits to simulate a pathogen attack and measure root exudation patterns as well as changes in associated microbial communities in response.
To accomplish this, we employ a reductionist approach, where microcosm systems are inoculated with defined synthetic bacterial communities and simulation of a pathogen attack is achieved by the application of elicitors on leaves. Furthermore, plants will be grown either on a defined clay substrate or in a hydroponics system developed for microbial manipulation and measuring of root exudates respectively.
First results show that members of the synthetic community associate with A. thaliana roots in both clay- and hydroponics systems at comparable densities. Furthermore, a shift in microbial composition between growth medium and roots could visually be assessed in both systems, indicating a selection of specific microbes at the root interface in the microcosms.
Next, we will determine the microbial community in the microcosms by 16S rRNA gene sequencing and/or selective plating. Additionally, we will finetune elicitor application on leaves to get a robust systemic immune stimulation. Finally, knowing that clay will absorb many compounds, we want to assess if it is suitable for root exudate sampling.
Break-out Room 4 chaired by Rie Inatsugi-Shimizu
17_Möhl, Patrick_Longer growing season: The dominant alpine sedge Carex curvula will not profit
Longer growing season: The dominant alpine sedge Carex curvula will not profit
Patrick Möhl1, Erika Hiltbrunner1
1 Department of Environmental Sciences, University of Basel
Temperate alpine grassland is adapted to a short growing season of a few months, constrained by cold temperature and snow cover. Ongoing climate warming has advanced snowmelt and confronts alpine plants with a longer growing season. This may prolong and enhance plant growth above- or belowground. Here, we assessed whether Carex curvula (the dominant alpine species on acidic soils in the Alps) is capable of sustaining growth and/or delay senescence when the season length is artificially extended by two to four months. Vegetation patches of alpine grassland were prematurely exposed to the growing season climate in a growth facility and compared to vegetation experiencing natural snowmelt in situ. Growth and senescence was quantified in C. curvula by measuring the length of the green part of individual leaves, which expands with growth and retracts during senescence. Autumn senescence in C.curvula started six to eight weeks after growth onset in spring, even under constant environmental conditions. Despite prolonging the growing season by up to 140%, senescence was slowed by up to 35% only. Accordingly, senescence considerably preceded the end of the growing season. Roots of the plant community were scanned using mini-rhizotrons with one transparent tube in every vegetation patch. Images were automatically segmented into roots and soil by means of machine learning, before the concentration of root pixels per image surface was extracted. Peak root growth occurred within the first two months, independent of season length and a longer growing season did not further stimulate root growth. In summary, our data suggests that growth and senescence of the dominant species C. curvula are strongly controlled by an internal clock that is tuned to the naturally occurring growing season length. A longer growing season under future climate change may therefore not benefit this species but promote others with a more flexible onset of autumn senescence.
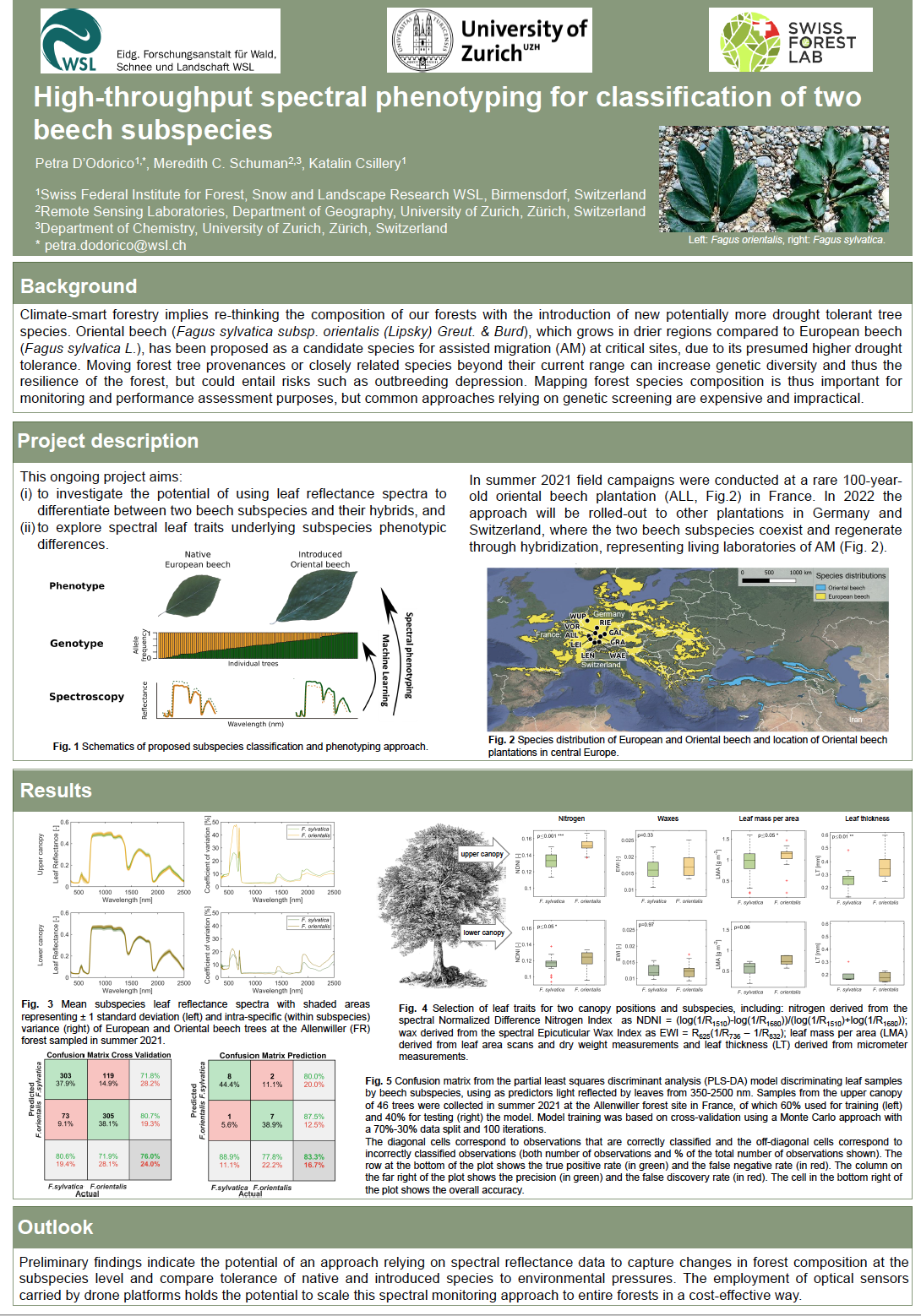
18_D’Odorico, Petra_High-throughput spectral phenotyping for classification of two beech subspecies
High-throughput spectral phenotyping for classification of two beech subspecies
Petra D’Odorico1, Meredith C. Schuman2,3, Katalin Csillery1
1 Swiss Federal Institute for Forest, Snow and Landscape Research WSL, Birmensdorf, Switzerland
2 Remote Sensing Laboratories, Department of Geography, University of Zurich
3 Department of Chemistry, University of Zurich
Climate-smart forestry implies re-thinking the composition of our forests with the introduction of new potentially more drought tolerant tree species. Oriental beech (Fagus sylvatica subsp. orientalis (Lipsky) Greut. & Burd), which grows in drier regions compared to European beech (Fagus sylvatica L.), has been proposed as a candidate species for assisted migration (AM) at critical sites, due to its presumed higher drought tolerance. Moving forest tree provenances or closely related species beyond their current range can increase genetic diversity and thus the resilience of the forest; but could entail risks as well, such as outbreeding depression.
Monitoring forest species composition is thus fundamental, but common genetic screening techniques are expensive and impractical. The recent advent of drones, as flexible platforms for remote sensing, has provided new opportunities for high-throughput phenotyping based on how light is reflected by leaves and crowns, and opened new opportunities for species characterization and separation.
This recently started project aims: (i) to investigate the potential of using leaf spectra to differentiate between two beech subspecies, and (ii) to understand the relationship between leaf spectral properties and the specific structural and chemical leaf traits underlying species differences. Taking advantage of a rare 100-year-old oriental beech plantation in France, representing a living laboratory of AM, where the two subspecies coexist and regenerate through hybridization, we conducted genotyping and phenotyping measurement campaigns. Phenotyping included the measurement of leaf and canopy spectra as well as determination of common structural and chemical leaf traits to be used in a machine learning model for species prediction.
Preliminary findings indicate the potential of an approach relying on spectral reflectance data to capture changes in forest species composition and compare tolerance of native and introduced species to environmental pressures. The advent of drones will allow scaling the spectral monitoring approach to entire tree populations in a cost-effective way.
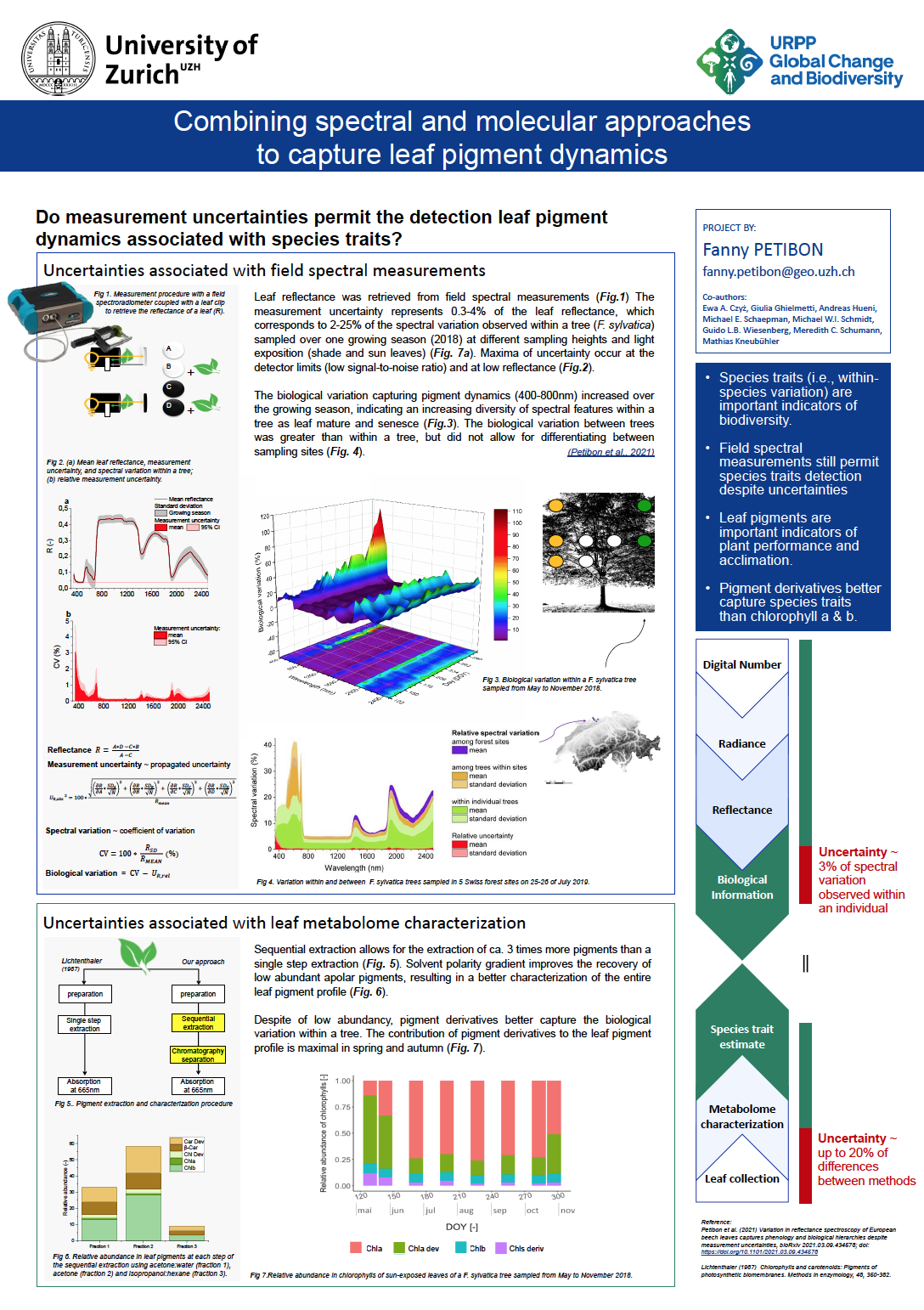
19_Petibon, Fanny_Combining spectral and molecular approaches to capture species trait variation
Combining spectral and molecular approaches to capture species trait variation
Fanny Petibon1, Ewa A. Czyż1, Giulia Ghielmetti1, Guido L.B. Wiesenberg1, Mathias Kneubühler1, Andreas Hueni1, Michael E. Schaepman1, Meredith C. Schuman1,2
1 Department of Geography - University of Zurich, Zurich, Switzerland
2 Department of Chemistry - University of Zurich, Zurich, Switzerland
Variation in trait values expressed by individuals result from a combination of biological and environmental factors. Such species trait variations are increasingly recognized as drivers and responses of biodiversity and ecosystem properties. However, little has been done to comprehensively characterize or monitor such variation using either spectral or molecular approaches, as emphasis is more often on species average values.
In this study we investigate what level of species trait variation in-situ leaf spectral measurements and leaf metabolomics can capture. We first developed a method to isolate species trait information from measurement uncertainty of leaf spectral measurements acquired with a field spectroradiometer coupled with a leaf clip. In parallel, we characterized complex leaf pigment profiles based on a newly developed liquid chromatography method, and epicuticular wax composition using a gas chromatography. We eventually compared the species trait variation observed within a Fagus sylvatica individual weekly sampled during the growing season 2018 to the variation observed among Fagus sylvatica individuals of a same population sampled in a Swiss forest.
We found that in-situ spectral measurements can detect species trait variation within a mature Fagus sylvatica tree sampled at various sampling positions and over time. Diversity of spectral features measured within an individual increased by 80% as leaves mature, with a contribution of measurement uncertainty of 3%. This increase correlated with an increasing diversity in pigment metabolites and changing wax composition. Besides, spectrally resolved species trait variation observed within a tree appeared to significantly contribute to the variation observed within a F. sylvatica population (up to 50%). We conclude that in-situ spectral measurements and leaf metabolomics have a great potential to better monitor and investigate species traits and full use has yet to be made of its potential.
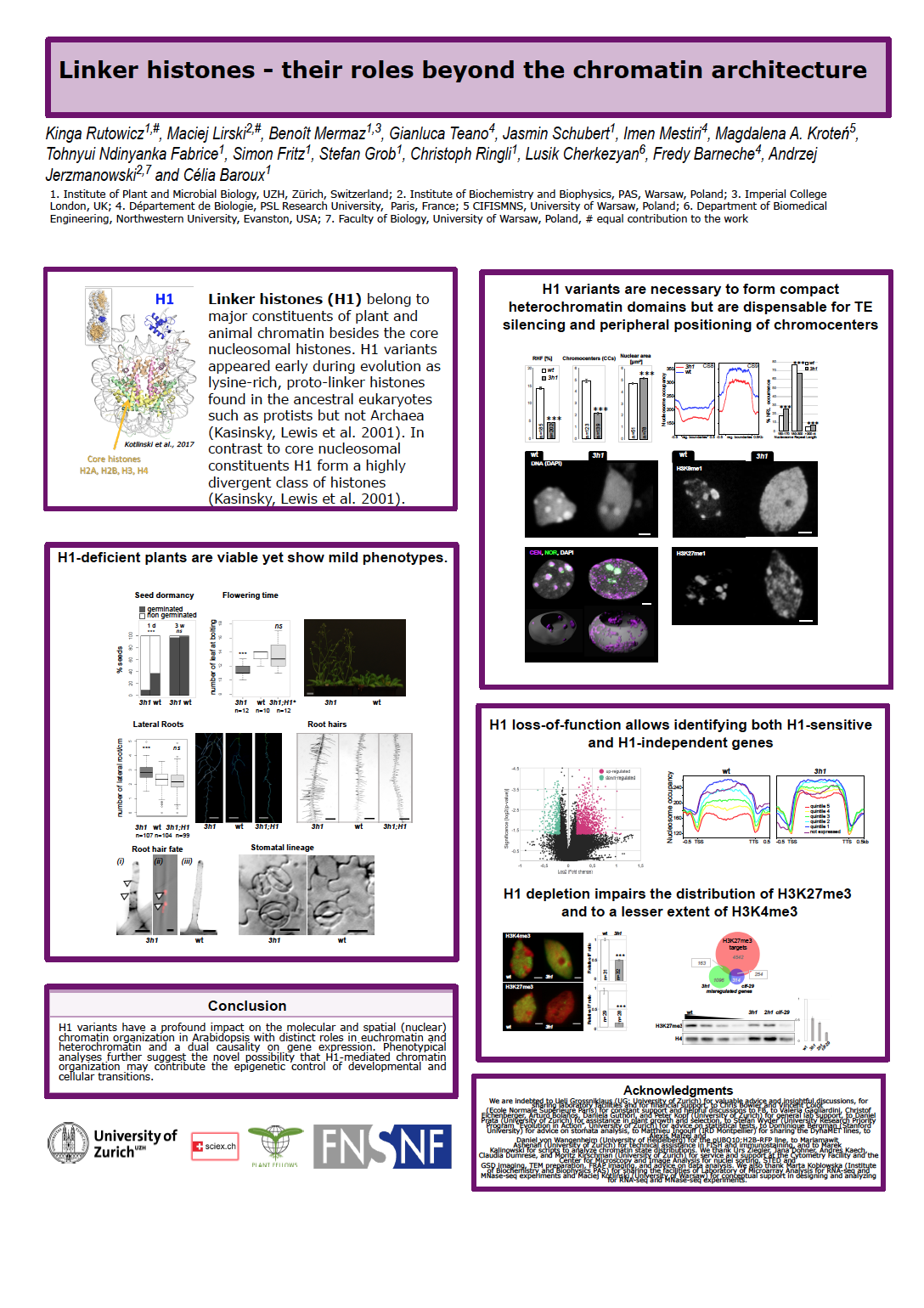
20_Rutowicz, Kinga_Linker histones - their roles beyond the chromatin architecture
Linker histones - their roles beyond the chromatin architecture
Kinga Rutowicz1, Maciej Lirski2, Benoît Mermaz1,6, Jasmin Schubert1, Gianluca Teano3, Imen Mestiri3, Magdalena A. Kroteń4, Fabrice Tonhyui1, Stefan Grob1, Christoph Ringli1, Lusik Cherkezeyan5, Fredy Barneche3, Andrzej Jerzmanowski2 and Célia Baroux1
1 Department of Plant and Microbial Biology, University of Zurich
2 Institute of Biochemistry and Biophysics, Polish Academy of Sciences, Warsaw, Poland
3 Département de Biologie, IBENS, Ecole Normale Supérieure, CNRS, INSERM, PSL Research University, Paris, France
4 College of Inter-Faculty Individual Studies in Mathematics and Natural Sciences, University of Warsaw, Poland
5 Department of Biomedical Engineering, Northwestern University, Evanston, USA
6 Department of Life Sciences, Imperial College London, South Kensington Campus, London, United-Kingdom
Chromatin provides a tunable platform for gene expression control. Besides the well-studied core nucleosome, H1 linker histones are abundant chromatin components with potential to influence chromatin function. Next to the intensively studied core histones which form the basic unit of chromatin – nucleosome - there is other abundant chromatin component histone H1. By combining the recent advances in microscopy and sequencing technics we showed not only that H1 is required for correct formation and organization of chromatin but also it is essential for maintaining proper chromatin decoration landscape, specifically H3 modifications. Lack of H1 in Arabidopsis plant did not severely affect the development yet it was associated with the failures in transitional fate changes in somatic cells e.g. lateral root formation, stomata patterning. Together, our data suggest that H1 fine tunes developmental transitions via regulating the precise chromatin organization and providing the accurate platform for chromatin modifying machinery and eventually epigenetics marks.
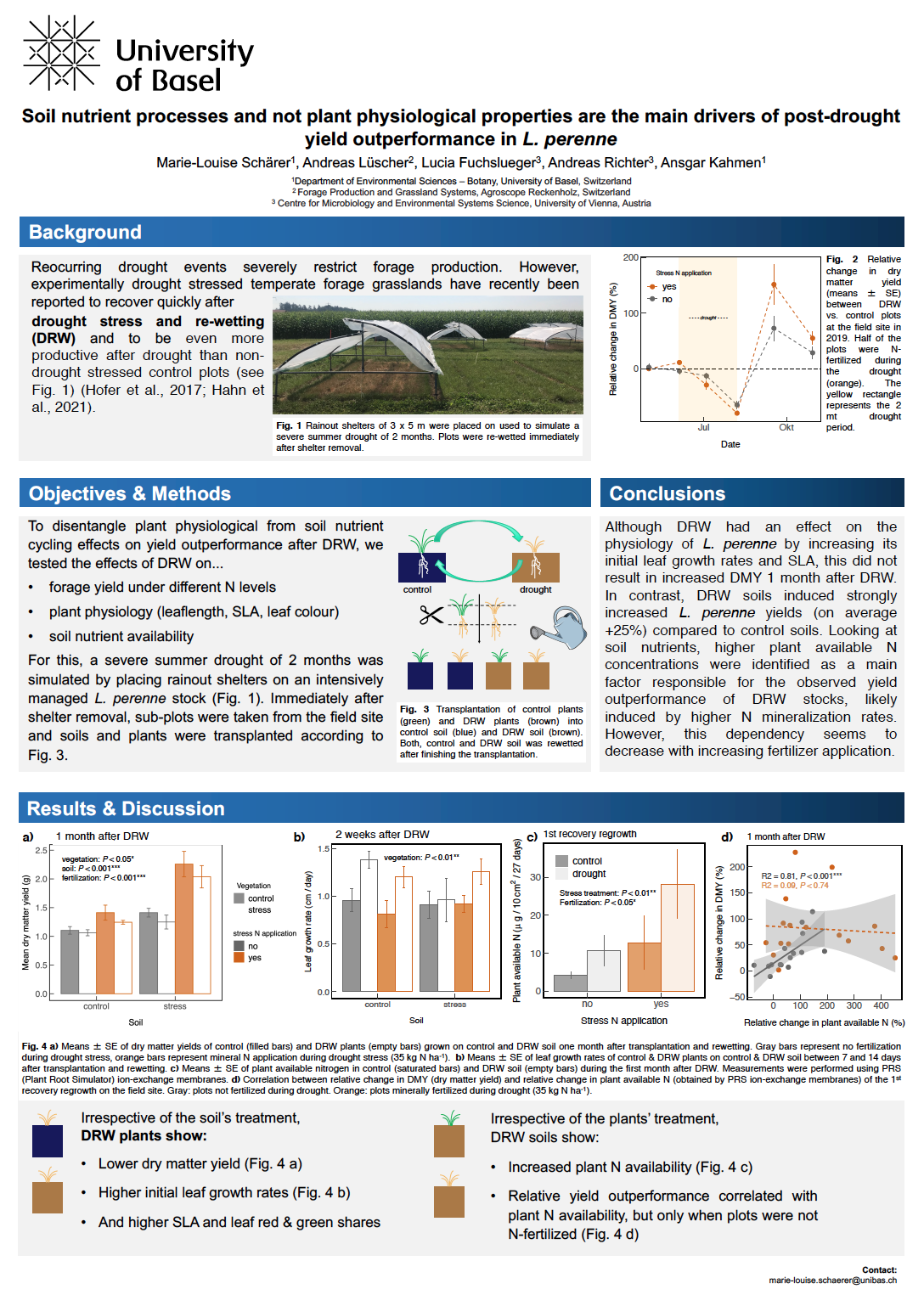
21_Schärer, Marie-Louise_Soil nutrient processes and not plant physiological properties are the main drivers of post-drought yield outperformance in L. perenne
Soil nutrient processes and not plant physiological properties are the main drivers of post-drought yield outperformance in L. perenne
Marie-Louise Schärer1, Andreas Lüscher2, Lucia Fuchslueger3, Andreas Richter3, Ansgar Kahmen1
1 Botanisches Institut, Universität Basel, Switzerland
2 Forage Production and Grassland Systems, Agroscope Reckenholz, Switzerland
3 Centre for Microbiology and Environmental Systems Science, University of Vienna, Austria
As we have seen in the past few years Reoccurring drought events can severely restrict forage production. A broad range of studies have examined the effect of drought on temperate forage grasslands. Experimentally drought stressed temperate forage grasslands recently been reported to recover quickly after drought stress and re-wetting (DRW) and to be even more productive after drought than non-drought stressed control plots (Hahn et al., 2021). However, an in-depth understanding of how plant physiological properties and soil nutrient cycling are affected by DRW and contribute to the outstanding post-drought yield outperformance is still missing. In this study we examined the effect of a 2-month experimental summer drought under different N availabilities on the recovery of a high-input L. perenne field stock. Concurrently, a post-drought transplantation experiment of control and DRW soil and plants withdrawn from the field was performed to disentangle plant physiological and soil nutrient cycling effects on yield recovery. Under all N conditions, DRW outperformed the control yield in the field. Irrespectively of the soils DRW treatment, transplanted DRW plants showed higher leaf growth rate and higher SLA. However, this did not result in higher dry matter yield of DRW plants. Instead, higher N mineralization rates and N and K availability in DRW soil were identified to be the main drivers of yield recovery in L. perenne.
Break-out Room 5 chaired by Klaus Schläppi
22_Stengele, Katja_Arabidopsis plant-soil feedbacks mediated by maize benzoxazinoids
Arabidopsis plant-soil feedbacks mediated by maize benzoxazinoids
Katja Stengele1, Henry Janse van Rensburg1, Klaus Schläppi1
1 Department of Environmental Sciences, University of Basel
Plants use root exudates to shape their immediate soil environment and assemble their root microbiota to improve their own fitness. These exudation-mediated changes in the soil microbiota can also be beneficial for the fitness of the second plant generation. Maize plants produce and secrete plant secondary metabolites called benzoxazinoids (BXs). Previously, Hu and colleagues [Nature Communications, 9, 2738 (2018)] showed that a second generation of wild-type maize displayed different growth and defense on soil previously conditioned by growing wild-type maize (BX+ soil) compared to soil conditioned with non-BX exuding bx1 mutant maize plants (BX– soil). Furthermore, the microbial community of the soil was differentially shaped between the two soil conditionings. However, if the BX-mediated soil conditioning also affects the growth of another, non-BX-producing plant species, is not well characterized. To address this gap, we tested the growth and defense feedbacks of the model plant Arabidopsis thaliana (Arabidopsis) on BX+ and BX– maize conditioned soils, and analysed the root microbiota compositions. Both the growth and defense responses of Arabidopsis were BX-conditioning dependent. Arabidopsis plants showed enhanced plant growth and resistance against the necrotrophic fungus Botrytis cinerea when grown on BX+ soil compared to BX– soil. Also, the bacterial community compositions of Arabidopsis roots differed between the two conditionings. These findings suggest that BX-mediated changes in soil bacterial communities translate to differences in the root bacterial communities, potentially explaining the growth and defense differences on BX-conditioned soils. However, the effect of different BX concentrations in the soil and the role of an interplay between the chemical (BX) and microbial soil components in modulating Arabidopsis feedbacks are not well understood so far. Thus, we aim to conduct continuing research based on this study system to further understand the growth and defense modulating effects of BX-conditioned soils.
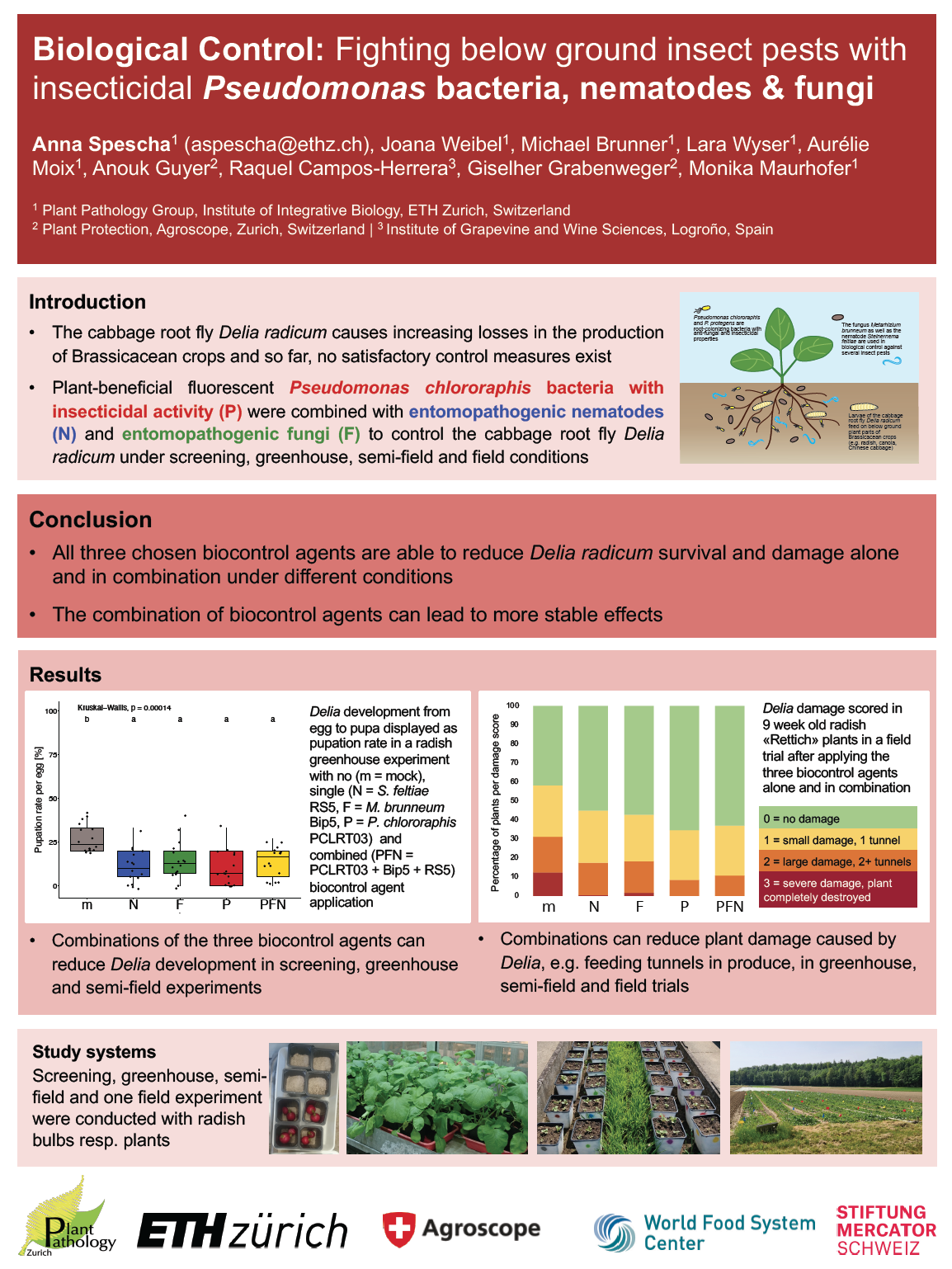
23_Spescha, Anna_Biological Control: Fighting below ground insect pests with entomopathogenic Pseudomonas bacteria, nematodes and fungi
Biological Control: Fighting below ground insect pests with entomopathogenic Pseudomonas bacteria, nematodes and fungi
Anna Spescha1, Michael Brunner1, Joana Weibel1, Franziska Scheibler1, Lara Wyser1, Aurélie Moix1, Florence Gillieron1, Mathias Hess1,2, Anouk Guyer2, Raquel Campos-Herrera3, Giselher Grabenweger2, Monika Maurhofer1
1 Institute of Integrative Biology, ETH Zurich, Switzerland
2 Plant Protection Unit, Agroscope, Zurich, Switzerland
3 Institute of Grapevine and Wine Sciences, Logroño, Spain
Below ground pests are difficult to control because either no effective control methods exist or suitable insecticides are or will be banned due to their negative effects on the soil and non-target organisms. Biological control is an environmental friendly alternative to insecticides. In this project, the combined application of soil and root inhabiting biocontrol agents as well as their interaction in the soil and in insects was investigated in various systems. First, different strains of entomopathogenic Pseudomonas bacteria, fungi and nematodes were screened in experiments against the cabbage root fly Delia radicum, an important pest in vegetable production. The most promising strains were subsequently upscaled and combined in greenhouse, semi-field and a field trial. Overall, the different biocontrol agents do not inhibit each other’s infectiousness and the combined application can even speed up the killing in the lab. Furthermore, the combined application lead to a reduced damage under semi-field and field conditions.
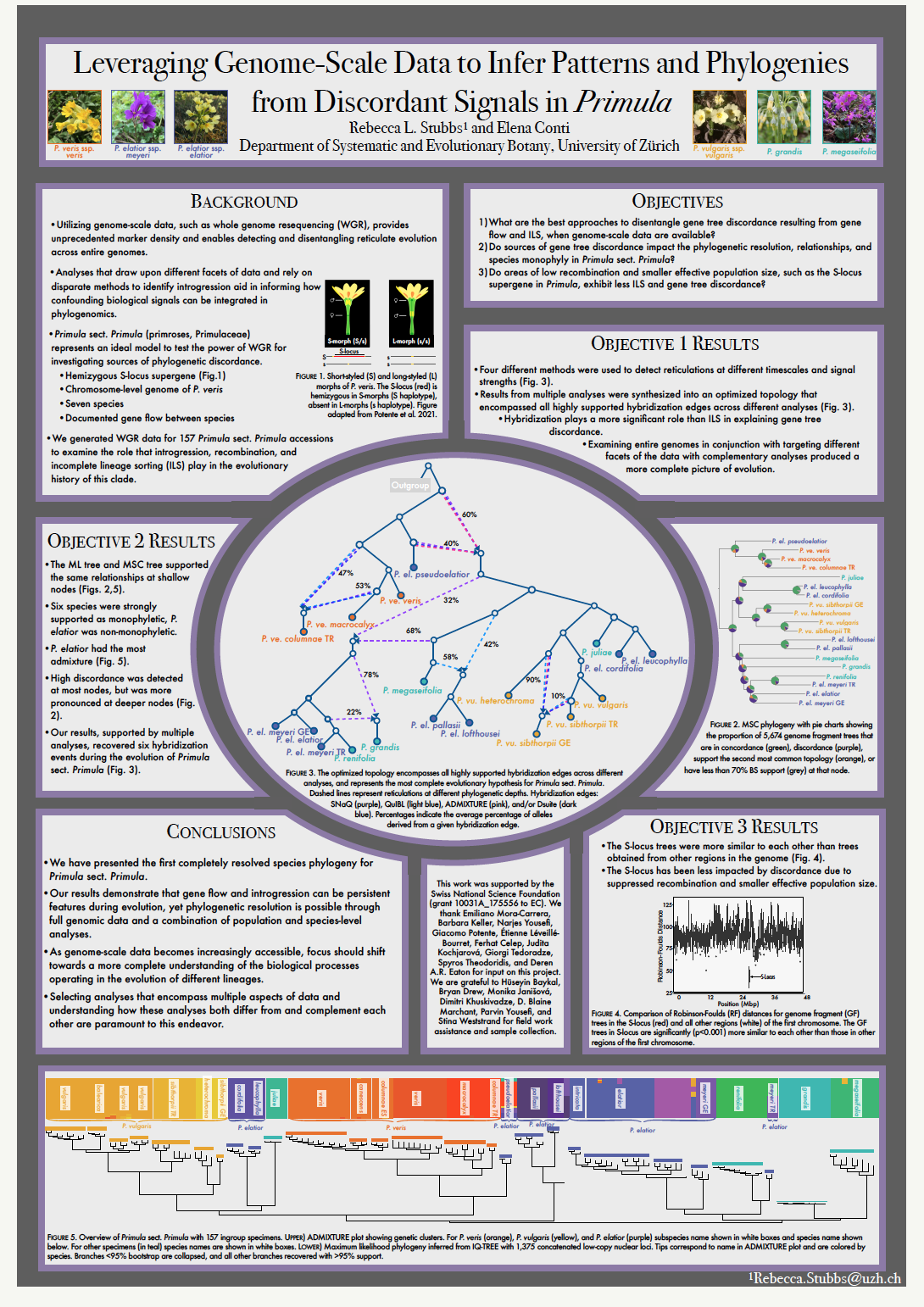
24_Stubbs, Rebecca_Leveraging Genome-Scale Data to Infer Patterns and Phylogenies from Discordant Signals
Leveraging Genome-Scale Data to Infer Patterns and Phylogenies from Discordant Signals
Rebecca Stubbs1, Elena Conti1
1 Department of Systematic and Evolutionary Botany, University of Zurich
Despite the advantages afforded by the vast increases in genomic data over the past decade, this profusion of data has been accompanied by an increase in theoretical and empirical complexities. This is in part due to larger data sets encompassing a wealth of signals across different genomic regions that often stray from a dichotomously branching phylogeny. These varied signals cover an array of both methodological complexities and biological processes at the level of genes and genomes, as well as individual organisms and populations. Additionally, underlying conflict stemming from processes such as recombination, hybridization, gene flow, and incomplete lineage sorting (ILS) can also confound phylogenetic inference. To explore the power of genome-scale data to provide resolution in groups with gene flow among species, we generated whole genome resequencing data for 160 specimens of Primula sect. Primula (Primulaceae). Through the use of this genomic data in conjunction with powerful analytical tools we dissect the causes of discordant genomic signals, thus enabling a more holistic understanding of species evolution through encompassing multiple evolutionary processes. Specifically, we explore how different analytical tools disentangle gene flow and ILS and what discordance reveals about the underlying evolutionary forces in this group of hybridizing plants.
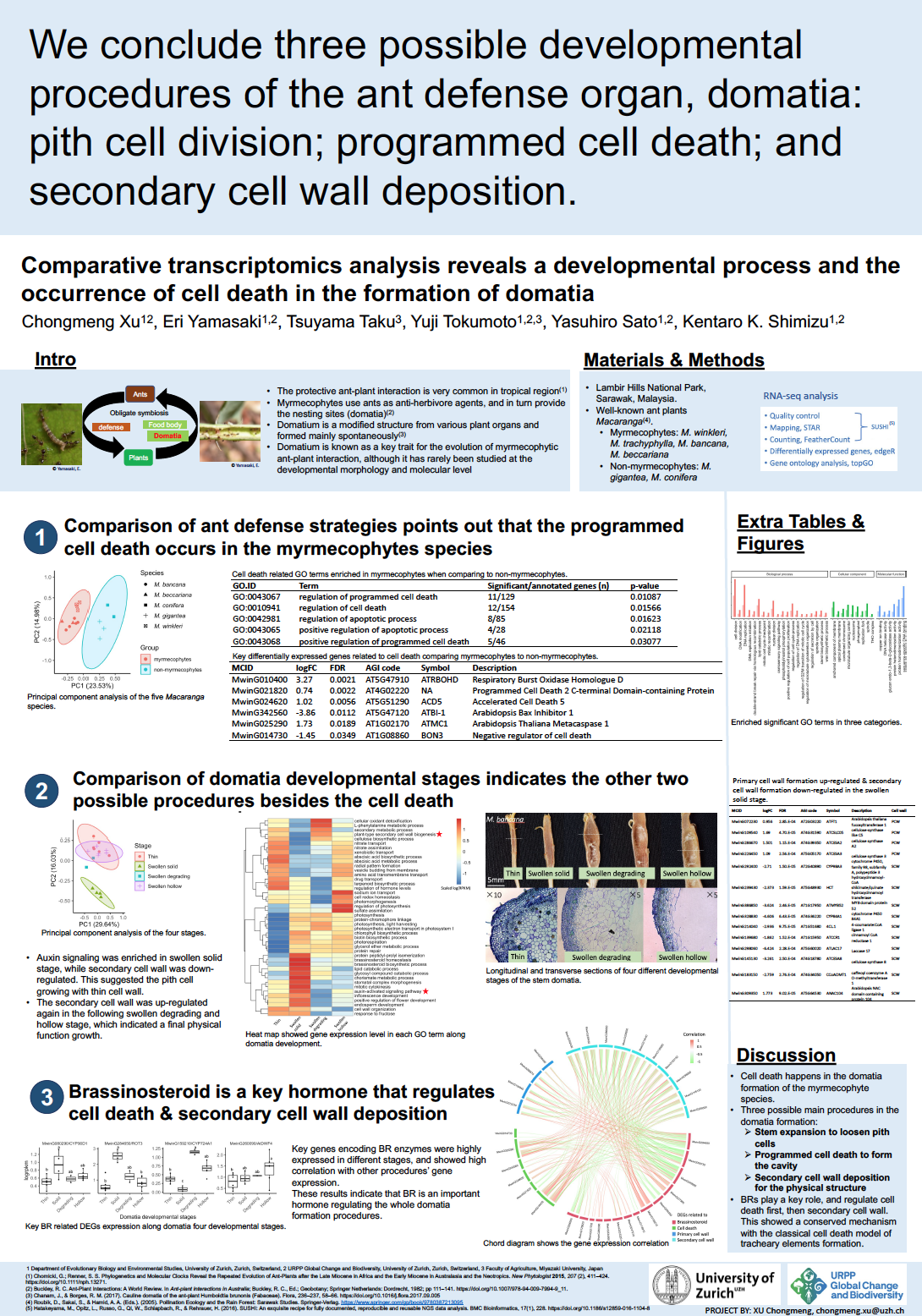
25_Xu, Chongmeng_Comparative transcriptomics analysis reveals a developmental process and the occurrence of cell death in the formation of domatia
Comparative transcriptomics analysis reveals a developmental process and the occurrence of cell death in the formation of domatia
Chongmeng Xu1, Eri Yamasaki1,2, Tsuyama Taku3, Yuji Tokumoto1,2,3, Yasuhiro Sato1,2, Kentaro K. Shimizu1,2
1 Department of Evolutionary Biology and Environmental Studies, University of Zurich
2 URPP Global Change and Biodiversity, University of Zurich
3 Faculty of Agriculture, Miyazaki University, Japan
Domatium is an important structure for obligate plant-ant mutualism, where myrmecophytes host, who use ants as anti-herbivore agents. Developmentally controlled programmed cell death (dPCD) is the ultimate step of cell specific differentiation in plants. Although domatia develop hollow cavities, no studies report the role of dPCD in their development. Even more, there are very few studies about the morphology and molecular mechanisms of the development of domatia. Comparative transcriptome and morphological analyses were used to study stem domatia formation among species that use different ant defense strategies and along the developmental stages within a species of the well-studied ant defense model genus Macaranga. PCD-related gene ontology (GO) terms and key differentially expressed genes (DEGs) were found in the species forming domatia whose pith cells were degrading. GO enrichment or DEGs highlighted the importance of brassinosteroids (BRs), primary cell wall (PCW) and secondary cell wall (SCW) during domatia development. These genetic results and morphological observations indicated that dPCD happened during the stem domatia formation. Furthermore, three possible procedures of domatia formation were pointed out: stem expansion to loosen pith cells; dPCD to form the cavity; and SCW deposition for the physical structure. In addition, it suggested that the BRs played a key role in regulating both PCD and SCW procedures to form the hollow domatia, which showed a conserved mechanism with the classical dPCD model of tracheary elements formation.
26_Zahnd, Cedric_Leaf phenology and non-structural carbohydrate dynamics along the vertical gradient of mature tree canopies
Leaf phenology and non-structural carbohydrate dynamics along the vertical gradient of mature tree canopies
Cedric Zahnd1, Lia Zehnder1, Ansgar Kahmen1, Günter Hoch1
1 Department of Environmental Sciences, University of Basel
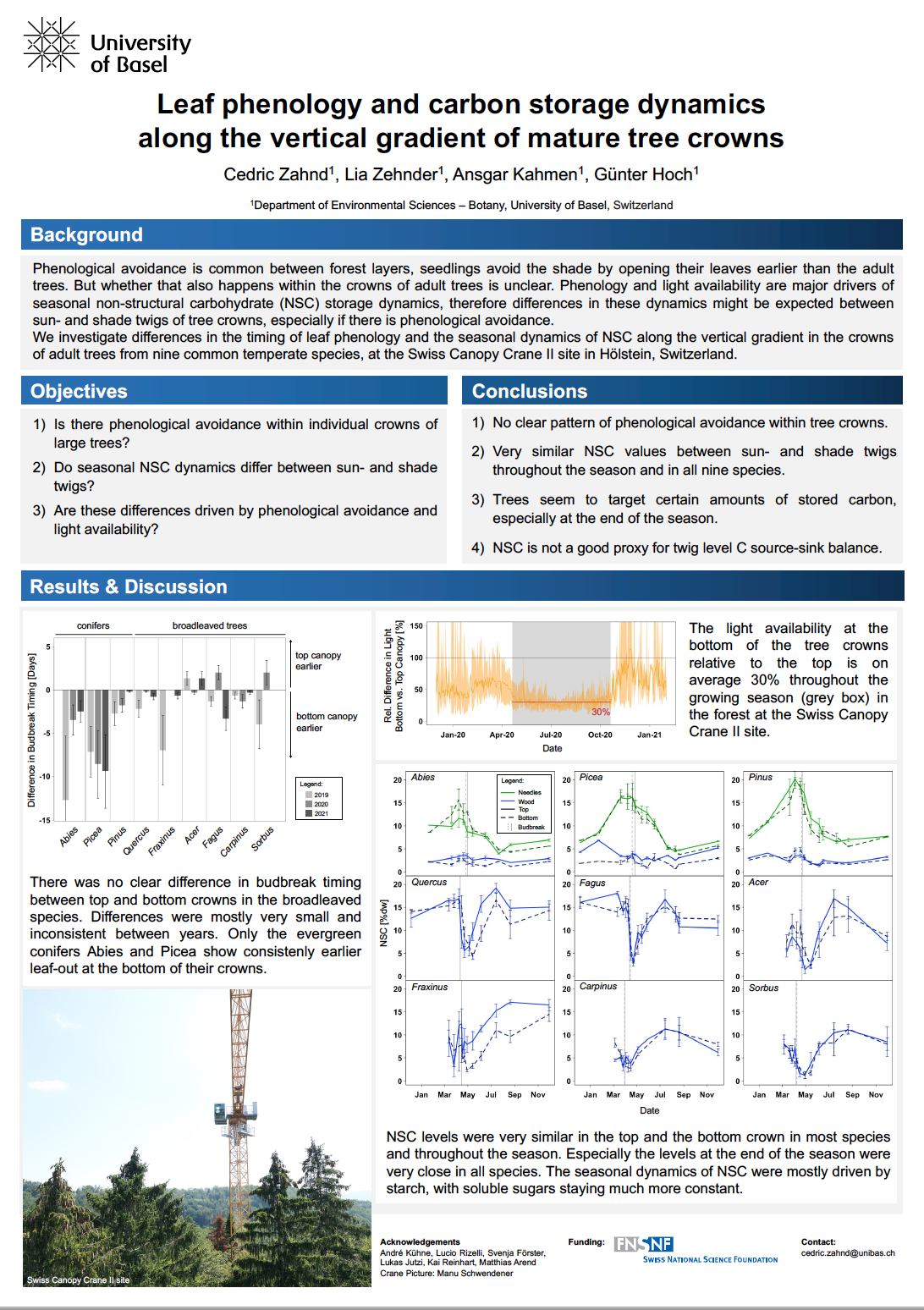
Light availability shows a strong gradient from the top to the bottom of tree crowns, leading to different amounts of carbon being sequestered along the depth of the canopy. With leaf phenology also potentially differing along that microclimatic gradient, it could be expected that the seasonal dynamics and size of the non-structural carbohydrate (NSC) pool of twigs would differ depending on their position within the crown.
To assess the effect of light availability and leaf phenology on the NSC dynamics along the vertical canopy gradient, we measured the NSC content in twigs from the top and bottom of the crowns of nine tree species in a mature, temperate forest near Basel, Switzerland, throughout the year 2020. Additionally, we recorded the leaf phenology along the vertical gradient of the crowns and took continuous light measurements at various locations within the canopy.
There was barely any difference in bud-break timing within the crowns, with the broadleaved trees showing bud-break on average only 1 day earlier at the bottom of the crown than at the top. The conifers showed more of a difference, with the bottom canopy having budbreak around 2 to 7 days earlier. Light availability in the lower crown was around 30% of that at the top throughout the growing season. In most species, the NSC concentrations were strikingly similar in top and bottom twigs throughout the season, maintaining the same NSC levels despite the stark differences in light availability. The only exceptions were the two ring-porous species Quercus petraea and Fraxinus excelsior, which both reached the minimum xylem starch levels about one week later at the bottom than the top. In both species, the lower crown was subsequently also delayed in refilling after leaf-out.
The very similar NSC concentrations throughout and especially at the end of the season support the idea of NSC storage being an actively regulated, rather than a passively driven pool.
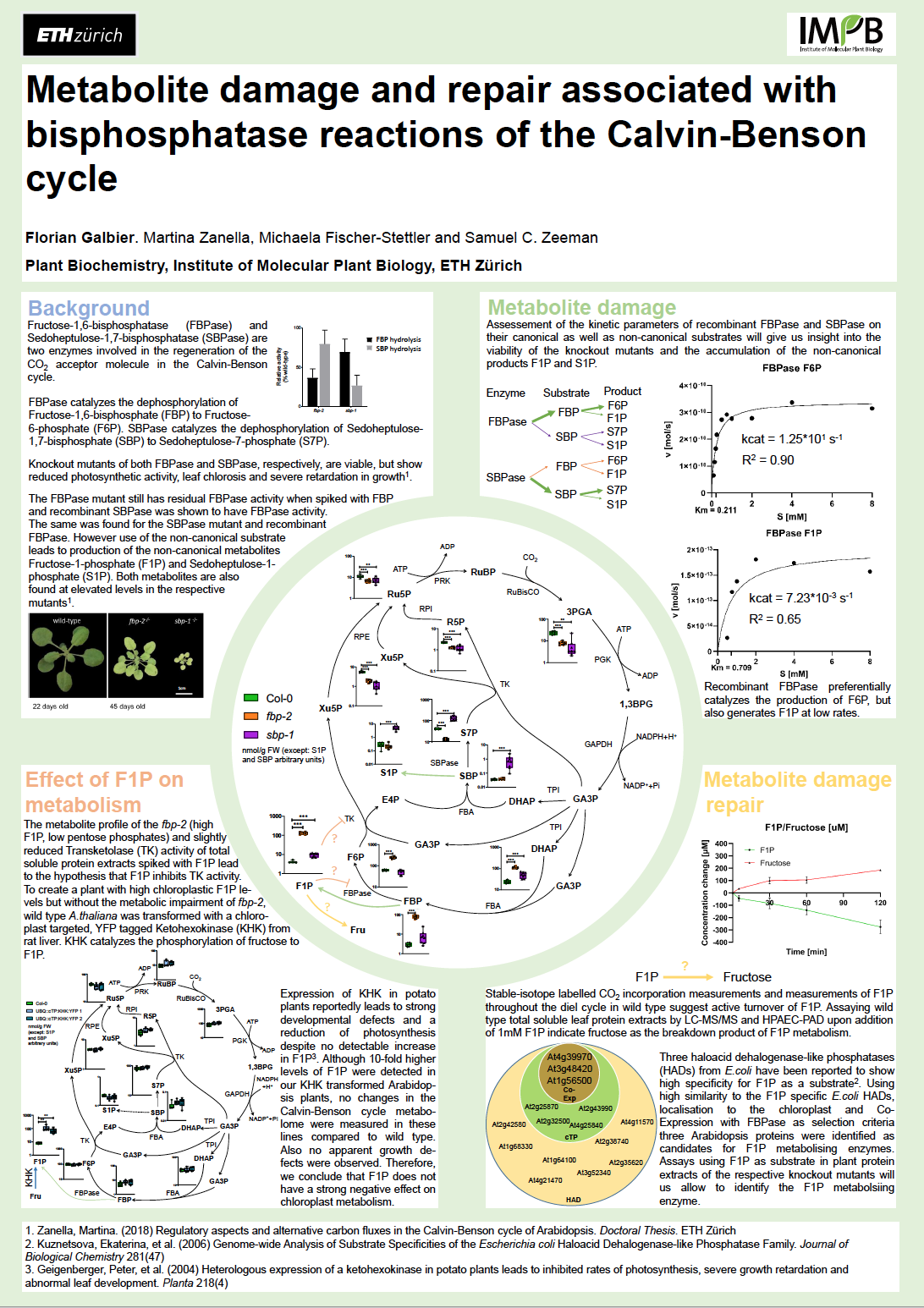
27_Galbier, Florian_Metabolite damage and repair associated with bisphosphatase reactions of the Calvin-Benson cycle
Metabolite damage and repair associated with bisphosphatase reactions of the Calvin-Benson cycle
Florian Galbier1, Martina Zanella1, Michaela Fischer-Stettler1, Samuel C. Zeeman1
1 Institute of Molecular Plant Biology, ETH Zurich
Metabolite damage and metabolite damage repair across all kingdoms of life receives increased attention. However, only very few of these pathways in plants have been elucidated. An example is the dephosphorylation of the RubisCO side reaction product xylulose-1,5-bisphosphate, a potent inhibitor of RubisCO, by the CbbY enzyme in Arabidopsis and Rhodobacter to the canonical Calvin-Benson cycle intermediate xylulose-5-phosphate. We describe new metabolite damage and repair pathways associated to the bisphosphatase reactions of the Calvin-Benson cycle - chloroplast fructose-1,6-bisphosphatase (FBPase), and sedoheptulose-1,7-bisphosphatase (SBPase). Chloroplast FBPase catalyzes the dephosphorylation of fructose-1,6-bisphosphate to fructose-6-phosphate (F6P), while SBPase catalyzes the dephosphorylation of sedoheptulose-1,7-bisphosphate to sedohepulose-7-phosphate. However, side reactions of both enzymes produce the 1-phosphate forms (i.e., F1P and S1P). Furthermore, both enzymes display substrate promiscuity, acting on the other’s canonical substrate and, when doing so, are more prone to catalyzing the side reaction. The substrate promiscuity renders Arabidopsis mutants in each enzyme viable but leads to accumulation of the side reaction product (i.e. F1P in the fbp1 mutant and S1P in the sbp mutant). We are exploring the repair pathways for the metabolism of F1P and S1P. In plant total soluble protein extracts F1P is metabolized to fructose, but the enzyme catalyzing this reaction remains to be identified. The resulting fructose could be reintegrated into the Calvin-Benson cycle by chloroplast fructokinase, yielding the canonical chloroplast FBPase product F6P. Other major pathways in central metabolism, such as glycolysis and the TCA cycle, have multiple metabolite damage and repair pathways associated with them and the same is highly likely to be true for the Calvin-Benson cycle. Our findings make a small contribution in unraveling the largely unknown metabolite damage and repair pathways associated with the Calvin-Benson cycle.